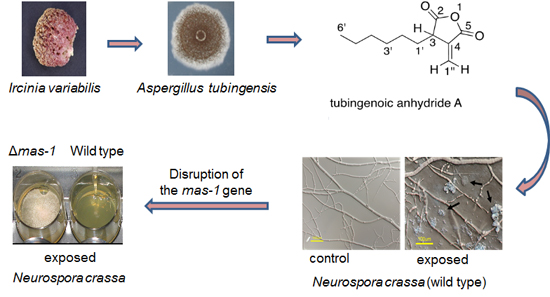
Sensitivity of Neurospora crassa to a Marine-Derived Aspergillus tubingensis Anhydride Exhibiting Antifungal Activity That Is Mediated by the MAS1 Protein
Abstract
:1. Introduction
2. Results and Discussion
2.1. A Marine-Derived Aspergillus tubingensis Produces Novel Antifungal Compounds


| Position | δC, Multiplicity | δH, Multiplicity | HMBC Correlations |
|---|---|---|---|
| 2 | 176.0, qC | H-3, H-1′a, H-1′b, H-1″a, H-1″b | |
| 3 | 46.6, CH | 3.40 t, 7.4 | H-1′a, H-1′b, H-1″a, H-1″b, H2-2′ |
| 4 | 138.5, qC | H-3, H-1′a, H-1′b, H-1″a, H-1″b | |
| 5 | 168.7, qC | H-3, H-1″a, H-1″b | |
| 1′ | 30.8, CH2 | a 1.82, ddt b 1.61, ddt | H-3, H2-2′, H2-3′ |
| 2′ | 27.3, CH2 | 1.25, m | H-3, H-1′a, H-1′b, H2-3′ |
| 3′ | 29.0, CH2 | 1.22, m | H-1′a, H-1′b, H2-2′ |
| 4′ | 31.4, CH2 | 1.22, m | H2-2′, H3-6′ |
| 5′ | 22.4, CH2 | 1.20, m | H2-3′, H3-6′ |
| 6′ | 13.8, CH3 | 0.82 t, 6.7 | H2-4′ |
| 1″ | 127.2, CH2 | a 5.71, s b 6.34, s | H-3 |
2.2. mas-1 Is a Novel N. crassa Gene Conferring Sensitivity to Antifungal Compounds Produced by A. tubingensis Strain OY907



2.3. Lack of mas-1 Increases the Resistance of N. crassa to Polyoxin D

2.4. Chitin Synthase Gene Expression Is Altered in Response to the A. tubingensis Extract

2.5. Cellular Localization of MAS1::GFP

3. Experimental Section
3.1. General Experimental Procedures—Chemical Analysis
3.2. Fungal Strains, Media, and Growth Conditions
3.3. Antifungal Activity Assays
3.4. Chemical Analysis of the Antifungal Product
 −6.3 (c 0.02, MeOH); UV (MeOH) λmax (log ɛ) 202 (3.58); IR (CHCl3) νmax 3020, 2929, 2858, 1708, 1629, 1522,1421,1212 cm−1; For NMR data see Table 1; HR ESI MS m/z 195.1024 ([M − H]−, calcd. for C11H15O3 m/z 195.1021).
−6.3 (c 0.02, MeOH); UV (MeOH) λmax (log ɛ) 202 (3.58); IR (CHCl3) νmax 3020, 2929, 2858, 1708, 1629, 1522,1421,1212 cm−1; For NMR data see Table 1; HR ESI MS m/z 195.1024 ([M − H]−, calcd. for C11H15O3 m/z 195.1021).3.5. Disruption of mas-1
3.6. Functional Complementation
3.7. Chitin Synthase Gene Expression
| Name | Sequence |
|---|---|
| f-1200 | CCAGTTCTTCTCCTCGTTCG |
| r-609 | TTCGTTCGCTGTCAATCAA |
| hphf | GTCGGAGACAGAAGATGATATTGAAGGAGC |
| hphr | GTTGGAGATTTCAGTAACGTTAAGTGGAT |
| mas-1f | ATGGCAAGGGCAAGAATTG |
| mas-1r | TCAGTGGTTGTGCCATTCAG |
| gfp-1f | AAGGTCTAGAATGGCAAGGG |
| gfp-1r | CCCGGGGTGGTTGTGCCATT |
| gfp-2f | GACGTAAACGGCCACAAGTT |
| gfp-2r | GAACTCCAGCAGGACCATGT |
| nat-1f | GGGCAGTAAGCGAAGGAGAATG |
| nat-1r | GGGATGGGAAGGATGGAGTA |
| chs-1f | GTGGGGTACCAAGGGTTCGG |
| chs-1r | CCTGTGGCTTCTCAATCTCT |
| chs-2f | TCTGGACAGCGACCTCAAGTTCAA |
| chs2-r | TGCCAAAGGCGTTGAAGAACCATC |
| chs-3f | TCAAGAACGATGTCGTCCAGCTCA |
| chs-3r | CAAAGGCCTGGAAGAACCAACGAT |
| chs-4f | TCTGGACTCGATTGCAATGACGGA |
| chs-4r | TTCCTCTCCGTGGCCCTTGATAAT |
| chs-5f | AAATCTCGGGCTTCTCATACGCCA |
| chs-5r | AACGGGCAGATGTGTCTTATCGCT |
| chs-6f | AAGACGGGTGACGACCTCAA |
| chs-6r | TAATGCCGGTGGTGAAGCCC |
| chs-7f | ACCCTCAACCTTACAGGCAACCTT |
| chs-7r | CAACAAGCCTTTGTCGGTGTCGAT |
3.8. MAS1::GFP Fusion Protein Construct and Confocal Imaging
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yarden, O. Fungal association with sessile marine invertebrates. Front. Microbiol. 2014, 5, 228. [Google Scholar] [CrossRef] [PubMed]
- Höller, U.; Wright, A.D.; Matthee, G.F.; Konig, G.M.; Draeger, S.; Aust, H.J.; Schulz, B. Fungi from marine sponges: Diversity, biological activity and secondary metabolites. Mycol. Res. 2000, 104, 1354–1365. [Google Scholar]
- Freeman, C.J.; Gleason, D.F. Chemical defenses, nutritional quality, and structural components in three sponge species: Ircinia felix, I. Campana, and Aplysina fulva. Mar. Biol. 2010, 157, 1083–1093. [Google Scholar]
- Kennedy, J.; Marchesi, J.R.; Dobson, A.D. Metagenomic approaches to exploit the biotechnological potential of the microbial consortia of marine sponges. Appl. Microbiol. Biotechnol. 2007, 75, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Siegl, A.; Hentschel, U. PKS and NRPS gene clusters from microbial symbiont cells of marine sponges by whole genome amplification. Environ. Microbiol. Rep. 2010, 2, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Rusch, D.; DeMaere, M.Z.; Yung, P.Y.; Lewis, M.; Halpern, A.; Heidelberg, K.B.; Egan, S.; Steinberg, P.D.; Kjelleberg, S. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010, 4, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Engel, S.; Pawlik, J.R. Allelopathic activities of sponge extracts. Mar. Ecol. Prog. Ser. 2000, 207, 273–281. [Google Scholar] [CrossRef]
- Compagnone, R.S.; Pina, I.C.; Rangel, H.R.; Dagger, F.; Suarez, A.I.; Reddy, M.V.R.; Faulkner, D.J. Antileishmanial cyclic peroxides from the palauan sponge Plakortis aff. Angulospiculatus. Tetrahedron 1998, 54, 3057–3068. [Google Scholar] [CrossRef]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Sabie, F.; Gadd, G. Effect of nucleosides and nucleotides and the relationship between cellular adenosine 3′:5′-cyclic monophosphate (cyclic amp) and germ tube formation in Candida albicans. Mycopathologia 1992, 119, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Abraham, W. Bioactive sesquiterpenes produced by fungi are they useful for humans as well. Curr. Med. Chem. 2001, 8, 583–606. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Shibashi, M. Bioactive metabolites of symbiotic marine microorganisms. Chem. Rev. 1993, 93, 1753–1769. [Google Scholar] [CrossRef]
- Lyons, P.C.; Plattner, R.D.; Bacon, C.W. Occurrence of peptide and clavine ergot alkaloids in tall fescue grass. Science 1986, 232, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Henning, D.; Schwarzer, M.; Marahiel, M. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 2002, 3, 490–504. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.M.; Jensen, P.R.; Fenical, W. A cyclic carbonate and related polyketides from a marine-derived fungus of the genus Phoma. Phytochemistry 2003, 64, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Ein-Gil, N.; Ilan, M.; Carmeli, S.; Smith, G.W.; Pawlik, J.R.; Yarden, O. Presence of Aspergillus sydowii, a pathogen of gorgonian sea fans in the marine sponge Spongia obscura. ISME J. 2009, 3, 752–755. [Google Scholar]
- Liu, H.B.; Edrada-Ebel, R.; Ebel, R.; Wang, Y.; Schulz, B.; Draeger, S.; Muller, W.E.G.; Wray, V.; Lin, W.H.; Proksch, P. Drimane sesquiterpenoids from the fungus Aspergillus ustus isolated from the marine sponge Suberites domuncula. J. Nat. Prod. 2009, 72, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Varoglu, M.; Crews, P. Biosynthetically diverse compounds from a saltwater culture of sponge-derived Aspergillus niger. J. Nat. Prod. 2000, 63, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Peng, C.S.; Ding, B.; Liu, F.; Zhang, F.L.; Lin, H.W.; Li, Z.Y. Biological active metabolite cyclo (l-trp-l-phe) produced by south china sea sponge Holoxea sp. associated fungus Aspergillus versicolor strain ts08. Bioprocess Biosyst. Eng. 2011, 34, 223–229. [Google Scholar] [CrossRef]
- Cohen, E.; Koch, L.; Thu, K.M.; Rahamim, Y.; Aluma, Y.; Ilan, M.; Yarden, O.; Carmeli, S. Novel terpenoids of the fungus Aspergillus insuetus isolated from the Mediterranean sponge Psammocinia sp. collected along the coast of Israel. Bioorgan. Med. Chem. 2011, 19, 6587–6593. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J. Biosynthesis of beta-glucans in fungi. Antonie Van Leeuwenhoek 1991, 60, 72–81. [Google Scholar]
- Osherov, N.; Yarden, O. The cell wall of filamentous fungi. In Cellular and Molecular Biology of Filamentous Fungi; Borkovich, K.A., Ebbole, D.J., Eds.; American Society for Microbiology: Washington, DC, USA, 2010; pp. 224–237. [Google Scholar]
- Endo, A.; Misato, T. Polyoxin D, a competitive inhibitor of UDP-N-acetylglucosamine: Chitin N-acetylglucosaminyltransferase in Neurospora crassa. Biochem. Biophysic. Res. Commun. 1969, 37, 718–722. [Google Scholar] [CrossRef]
- Muller, H.; Furter, R.; Zahner, H.; Rast, D.M. Metabolic products of microorganisms. Inhibition of chitosomal chitin synthetase and growth of Mucor-rouxii by Nikkomycin-Z, Nikkomycin-X, and Polyoxin-A: A comparison. Arch. Microbiol. 1981, 130, 195–197. [Google Scholar]
- Beauvais, A.; Latge, J.P. Membrane and cell wall targets in Aspergillus fumigatus. Drug Resist. Updat. 2001, 4, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Bruneau, J.M.; Mol, P.C.; Buitrago, M.J.; Legrand, R.; Latge, J.P. Glucan synthase complex of Aspergillus fumigatus. J. Bacteriol. 2001, 183, 2273–2279. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W. Echinocandin antifungal drugs. Lancet 2003, 362, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Talontsi, F.M.; Tatong, M.D.; Dittrich, B.; Douanla-Meli, C.; Laatsch, H. Structures and absolute configuration of three alpha-pyrones from an endophytic fungus Aspergillus niger. Tetrahedron 2013, 69, 7147–7151. [Google Scholar] [CrossRef]
- Paz, Z.; Komon-Zelazowska, M.; Druzhinina, I.S.; Aveskamp, M.M.; Shnaiderman, A.; Aluma, Y.; Carmeli, S.; Ilan, M.; Yarden, O. Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 2010, 42, 17–26. [Google Scholar] [CrossRef]
- Samson, R.A.; Seifert, K.A.; Kuijpers, A.F.; Houbraken, J.A.; Frisvad, J.C. Phylogenetic analysis of Penicillium subgenus Penicillium using partial β-tubulin sequences. Stud. Mycol. 2004, 49, 175–200. [Google Scholar]
- Thirunavukkarasu, N.; Suryanarayanan, T.S.; Girivasan, K.P.; Venkatachalam, A.; Geetha, V.; Ravishankar, J.P.; Doble, M. Fungal symbionts of marine sponges from Rameswaram, southern India: Species composition and bioactive metabolites. Fungal Divers. 2012, 55, 37–46. [Google Scholar] [CrossRef]
- Ding, B.; Yin, Y.; Zhang, F.L.; Li, Z.Y. Recovery and phylogenetic diversity of culturable fungi associated with marine sponges Clathrina luteoculcitella and Hloxea sp. in the South China Sea. Mar. Biotechnol. 2011, 13, 713–721. [Google Scholar] [CrossRef]
- Weidenmuller, H.L.; Cavagna, G.; Fehlhaber, H.W.; Prave, P. 2-carboxymethyl-3-hexylmaleic acid anhydride, A novel metabolite from an Aspergillus. Tetrahedron Lett. 1972, 33, 3519–3533. [Google Scholar] [CrossRef]
- Mondal, G.; Dureja, P.; Sen, B. Fungal metabolites from Aspergillus niger AN27 related to plant growth promotion. Indian J Exp. Biol. 2000, 38, 84–87. [Google Scholar] [PubMed]
- Keogh, M.F.; Zurita, M.E. α-(15-hydroxyhexadecyl) itaconic acid from Usnea awphatica. Phytochemistry 1977, 16, 134–135. [Google Scholar] [CrossRef]
- Buttery, R.G.; Seifert, R.M.; Haddon, W.F.; Lundin, R.E. 2-Hexyl-3-methylmaleic anhydride: An unusual volatile component of raisins and almond hulls. J. Agric. Food Chem. 1980, 28, 1336–1338. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Borkovich, K.A.; Selker, E.U.; Read, N.D.; Jaffe, D.; FitzHugh, W.; Ma, L.J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Borkovich, K.A.; Alex, L.A.; Yarden, O.; Freitag, M.; Turner, G.E.; Read, N.D.; Seiler, S.; Bell-Pedersen, D.; Paietta, J.; Plesofsky, N.; et al. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004, 68, 1–108. [Google Scholar] [CrossRef] [PubMed]
- Staben, C.; Jensen, B.; Singer, M.; Pollock, J.; Schechtman, M.; Kinsey, J.; Selker, E. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newslett. 1989, 36, 79–81. [Google Scholar]
- Carrillo-Munoz, A.J.; Giusiano, G.; Ezkurra, P.A.; Quindos, G. Antifungal agents: Mode of action in yeast cells. Rev. Esp. Quimioter. 2006, 19, 130–139. [Google Scholar]
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Beth Din, A.B.; Specht, C.A.; Robbins, P.W.; Yarden, O. chs-4, a class IV chitin synthase gene from Neurospora crassa. Mol. Gen. Genet. 1996, 250, 214–222. [Google Scholar]
- Freitag, M.; Hickey, P.C.; Raju, N.B.; Selker, E.U.; Read, N.D. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 2004, 41, 897–910. [Google Scholar] [CrossRef] [PubMed]
- Sietsma, J.H.; Din, A.B.; Ziv, V.; Sjollema, K.A.; Yarden, O. The localization of chitin synthase in membranous vesicles (chitosomes) in Neurospora crassa. Microbiology 1996, 142, 1591–1596. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.; Bartnicki-Garcia, S.; Gonzalez-Prieto, J.M.; Sanchez-Leon, E.; Verdin-Ramos, J.A.; Beltran-Aguilar, A.; Freitag, M. Spitzenkörper localization and intracellular traffic of green fluorescent protein-labeled chs-3 and chs-6 chitin synthases in living hyphae of Neurospora crassa. Eukaryot. Cell 2007, 6, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Beth Din, A.; Yarden, O. The Neurospora crassa chs-2 gene encodes a non-essential chitin synthase. Microbiology 1994, 140, 2189–2197. [Google Scholar]
- Davis, R.H. Neurospora—Contributions of a Model Organism; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Morton, D.J.; Stroube, W.H. Antagonistic and stimulatory effects of soil microorganisms upon Sclerotium rolfsii. Phytopathology 1955, 45, 417–420. [Google Scholar]
- Ziv, C.; Yarden, O. Gene silencing for functional analysis: Assessing RNAi as a tool for manipulation of gene expression. Mol. Cell Biol. Methods Fungi 2010, 638, 77–100. [Google Scholar]
- Seiler, S.; Vogt, N.; Ziv, C.; Gorovits, R.; Yarden, O. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol. Biol. Cell 2006, 17, 4080–4092. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, S.; Freeman, S.; Zveibil, A.; Yarden, O. A defect in nir1, a nirA-like transcription factor, confers morphological abnormalities and loss of pathogenicity in Colletotrichum acutatum. Mol. Plant Pathol. 2006, 7, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Salame, T.M.; Yarden, O.; Hadar, Y. Pleurotus ostreatus manganese-dependent peroxidase silencing impairs decolourization of Orange II. Microb. Biotechnol. 2010, 3, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- McNally, M.T.; Free, S.J. Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 1988, 14, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Gorovits, R.; Yarden, O. Environmental suppression of Neurospora crassa COT1 hyperbranching: A link between COT1 kinase and stress sensing. Eukaryot. Cell 2003, 2, 699–707. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Koch, L.; Lodin, A.; Herold, I.; Ilan, M.; Carmeli, S.; Yarden, O. Sensitivity of Neurospora crassa to a Marine-Derived Aspergillus tubingensis Anhydride Exhibiting Antifungal Activity That Is Mediated by the MAS1 Protein. Mar. Drugs 2014, 12, 4713-4731. https://doi.org/10.3390/md12094713
Koch L, Lodin A, Herold I, Ilan M, Carmeli S, Yarden O. Sensitivity of Neurospora crassa to a Marine-Derived Aspergillus tubingensis Anhydride Exhibiting Antifungal Activity That Is Mediated by the MAS1 Protein. Marine Drugs. 2014; 12(9):4713-4731. https://doi.org/10.3390/md12094713
Chicago/Turabian StyleKoch, Liat, Anat Lodin, Inbal Herold, Micha Ilan, Shmuel Carmeli, and Oded Yarden. 2014. "Sensitivity of Neurospora crassa to a Marine-Derived Aspergillus tubingensis Anhydride Exhibiting Antifungal Activity That Is Mediated by the MAS1 Protein" Marine Drugs 12, no. 9: 4713-4731. https://doi.org/10.3390/md12094713
APA StyleKoch, L., Lodin, A., Herold, I., Ilan, M., Carmeli, S., & Yarden, O. (2014). Sensitivity of Neurospora crassa to a Marine-Derived Aspergillus tubingensis Anhydride Exhibiting Antifungal Activity That Is Mediated by the MAS1 Protein. Marine Drugs, 12(9), 4713-4731. https://doi.org/10.3390/md12094713