Identification of a 4-Deoxy-l-erythro-5-hexoseulose Uronic Acid Reductase, FlRed, in an Alginolytic Bacterium Flavobacterium sp. Strain UMI-01
Abstract
:1. Introduction
2. Results
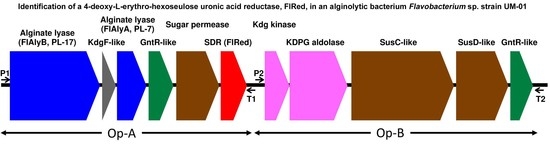
2.1. Identification of FlRed Gene in Strain UMI-01 Genome


2.2. Enzymatic Properties of Recombinant FlRed

| Substrates | Specific Activity (U/mg) | Relative Activity (%) |
|---|---|---|
| Aldehydes | ||
| DEH | ||
| (NADH) | 4.0 ± 0.28 | 100 |
| (NADPH) | 0.043 ± 0.002 | 1.1 |
| Benzaldehyde | N.D | N.D |
| Glutaraldehyde | N.D | N.D |
| o-Phthalaldehyde | N.D | N.D |
| Ketones | ||
| 4-Methyl-2-pentanone | N.D | N.D |
| 2,5-Hexanedione | N.D | N.D |
| 3-Chloropropiophenone | N.D | N.D |
| Ethylbenzoylacetate | N.D | N.D |
| Keto ester | ||
| Methyl pyruvate | N.D | N.D |
| α-Keto acid | ||
| α-Keto-glutaric acid | N.D | N.D |
| Aldose | ||
| Glucose | N.D | N.D |
| Galactose | N.D | N.D |


3. Discussion
4. Experimental Section
4.1. Materials
4.2. Genome Analysis for Flavobacterium sp. Strain UMI-01
4.3. Production of Recombinant FlRed

4.4. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
4.5. Determination of Protein Concentration
4.6. Assay for Alginate Lyase Activity
4.7. Preparation of Crude Extract from Strain UMI-01

4.8. Preparation of DEH
4.9. Assay for DEH-Reducing Activity
4.10. Assessment of Substrate Specificity of RecFlred
4.11. Thin-Layer Chromatography of Reaction Products
4.12. Mass Spectrometry for DEH and KDG
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Haug, A.; Larsen, B.; Smidsrod, O. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem. Scand. 1967, 21, 691–704. [Google Scholar] [CrossRef]
- Gacesa, P. Alginates. Carbohydr. Polym. 1988, 8, 161–182. [Google Scholar] [CrossRef]
- Gacesa, P. Enzymatic degradation of alginates. Int. J. Biochem. 1992, 24, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Tomoda, Y.; Umemura, K.; Adachi, T. Promotion of barley root elongation under hypoxic conditions by alginate lyase-lysate. Biosci. Biotechnol. Biochem. 1994, 58, 202–203. [Google Scholar] [CrossRef]
- Sutherland, I.W. Polysaccharide lyases. FEMS Microbiol. Rev. 1995, 16, 323–347. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Iwamoto, Y.; Kitamura, Y.; Oda, T.; Muramatsu, T. Root growth-promoting activity of unsaturated oligomericuronates from alginate on carrot and rice plants. Biosci. Biotechnol. Biochem. 2003, 67, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Endo, T.; Nakakita, R.; Murata, K.; Yonemoto, Y.; Okayama, K. Effect of depolymerized alginates on the growth of Bifidobacteria. Biosci. Biotechnol. Biochem. 1992, 56, 355–356. [Google Scholar] [CrossRef] [PubMed]
- Ariyo, B.; Tamerler, C.; Bucke, C.; Keshavarz, T. Enhanced penicillin production by oligosaccharides from batch culture of Penicillium chrysogenum in stirred-tank reactors. FEMS Microbiol. Lett. 1998, 166, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Kawada, A.; Hiura, N.; Tajima, S.; Takahara, H. Alginate oligosaccharides stimulate VEGF-mediated growth and migration of human endothelial cells. Arch. Dermatol. Res. 1999, 291, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Chaki, T.; Ogawa, H. Effect of sodium alginate oligosaccharide on human blood pressure. Med. Cons. New-Remed. 2001, 38, 555–560. (In Japanese) [Google Scholar]
- Chaki, T.; Nishimoto, S.; Hiura, N.; Satou, R.; Tou, Y.; Kakinuma, S. Effect of a powdered drink containing sodium alginate oligosaccharide on blood pressure in mild hypertensive and high normal blood pressure subjects. J. Nutr. Food 2002, 5, 41–54. (In Japanese) [Google Scholar]
- Takeda, H.; Yoneyama, F.; Kawai, S.; Hashimoto, W.; Murata, K. Bioethanol production from marine biomass alginate by metabolically engineered bacteria. Energy Environ. Sci. 2011, 4, 2575–2581. [Google Scholar] [CrossRef]
- Wargacki, A.J.; Leonard, E.; Win, M.N.; Regitsky, D.D.; Santos, C.N.; Kim, P.B.; Cooper, S.R.; Raisner, R.M.; Herman, A.; Sivitz, A.B.; et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 2012, 335, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Enquist-Newman, M.; Faust, A.M.; Bravo, D.D.; Santos, C.N.; Raisner, R.M.; Hanel, A.; Sarvabhowman, P.; Le, C.; Regitsky, D.D.; Cooper, S.R.; et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 2014, 505, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J.; Ashwell, G. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 1962, 237, 309–316. [Google Scholar]
- Preiss, J.; Ashwell, G. Alginic acid metabolism in bacteria. II. The enzymatic reduction of 4-deoxy-l-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-d-gluconic acid. J. Biol. Chem. 1962, 237, 317–321. [Google Scholar] [PubMed]
- Takase, R.; Ochiai, A.; Mikami, B.; Hashimoto, W.; Murata, K. Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochim. Biophys. Acta 2010, 1804, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Persson, B.; Kallberg, Y. Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs). Chem. Biol. Interact. 2013, 202, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Takadono, K.; Nishiyama, R.; Tajima, K.; Kobayashi, T.; Ojima, T. Characterization of an alginate lyase, Flalya, from Flavobacterium sp. strain UMI-01 and its expression in Escherichia coli. Mar. Drugs 2014, 12, 4693–4712. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Bagdasarian, M.; Kaufman, M.G.; Walker, E.D. Characterization of strong promoters from an environmental Flavobacterium hibernum strain by using a green fluorescent protein-based reporter system. Appl. Environ. Microbiol. 2007, 73, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Bagdasarian, M.; Kaufman, M.G.; Bates, A.K.; Walker, E.D. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J. Bacteriol. 2007, 189, 5108–5118. [Google Scholar] [CrossRef] [PubMed]
- Nasser, W.; Reverchon, S.; Condemine, G.; Robert-Baudouy, J. Specific interactions of Erwinia chrysanthemi KdgR repressor with different operators of genes involved in pectinolysis. J. Mol. Biol. 1994, 236, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.J.; Hahnke, R.L.; Huang, S.; Werner, J.; Xing, P.; Barbeyron, T.; Huettel, B.; Stüber, K.; Reinhardt, R.; Harder, J.; et al. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl. Environ. Microbiol. 2013, 79, 6813–6822. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Lundqvist, L.C.; Jam, M.; Jeudy, A.; Barbeyron, T.; Sandström, C.; Michel, G.; Czjzek, M. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 2013, 288, 23021–23037. [Google Scholar] [CrossRef] [PubMed]
- Pati, A.; Abt, B.; Teshima, H.; Nolan, M.; Lapidus, A.; Lucas, S.; Hammon, N.; Deshpande, S.; Cheng, J.F.; Tapia, R.; et al. Complete genome sequence of Cellulophaga lytica type strain (LIM-21). Stand. Genomic Sci. 2011, 4, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Starns, D.; Oshima, K.; Suda, W.; Iino, T.; Yuki, M.; Inoue, J.; Kitamura, K.; Iida, T.; Darby, A.; Hattori, M.; et al. Draft genome sequence of Cytophaga fermentans JCM 21142T, a facultative anaerobe isolated from marine mud. Genome Announc. 2014, 2, e00206–e00214. [Google Scholar] [CrossRef] [PubMed]
- Filling, C.; Berndt, K.D.; Benach, J.; Knapp, S.; Prozorovski, T.; Nordling, E.; Ladenstein, R.; Jörnvall, H.; Oppermann, U. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 2002, 277, 25677–25684. [Google Scholar] [CrossRef] [PubMed]
- Takase, R.; Mikami, B.; Kawai, S.; Murata, K.; Hashimoto, W. Structure-based conversion of the coenzyme requirement of a short-chain dehydrogenase/reductase involved in bacterial alginate metabolism. J. Biol. Chem. 2014, 289, 33198–33214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porzio, M.A.; Pearson, A.M. Improved resolution of myofibrillar proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochim. Biophys. Acta 1977, 490, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Gornall, A.G.; Bardawill, C.J.; David, M.M. Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 1949, 177, 751–766. [Google Scholar] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Lin, F.M.; Pomeranz, Y. Effect of borate on colorimetric determinations of carbohydrates by the phenol-sulfuric acid method. Anal. Biochem. 1968, 24, 128–131. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inoue, A.; Nishiyama, R.; Mochizuki, S.; Ojima, T. Identification of a 4-Deoxy-l-erythro-5-hexoseulose Uronic Acid Reductase, FlRed, in an Alginolytic Bacterium Flavobacterium sp. Strain UMI-01. Mar. Drugs 2015, 13, 493-508. https://doi.org/10.3390/md13010493
Inoue A, Nishiyama R, Mochizuki S, Ojima T. Identification of a 4-Deoxy-l-erythro-5-hexoseulose Uronic Acid Reductase, FlRed, in an Alginolytic Bacterium Flavobacterium sp. Strain UMI-01. Marine Drugs. 2015; 13(1):493-508. https://doi.org/10.3390/md13010493
Chicago/Turabian StyleInoue, Akira, Ryuji Nishiyama, Shogo Mochizuki, and Takao Ojima. 2015. "Identification of a 4-Deoxy-l-erythro-5-hexoseulose Uronic Acid Reductase, FlRed, in an Alginolytic Bacterium Flavobacterium sp. Strain UMI-01" Marine Drugs 13, no. 1: 493-508. https://doi.org/10.3390/md13010493
APA StyleInoue, A., Nishiyama, R., Mochizuki, S., & Ojima, T. (2015). Identification of a 4-Deoxy-l-erythro-5-hexoseulose Uronic Acid Reductase, FlRed, in an Alginolytic Bacterium Flavobacterium sp. Strain UMI-01. Marine Drugs, 13(1), 493-508. https://doi.org/10.3390/md13010493