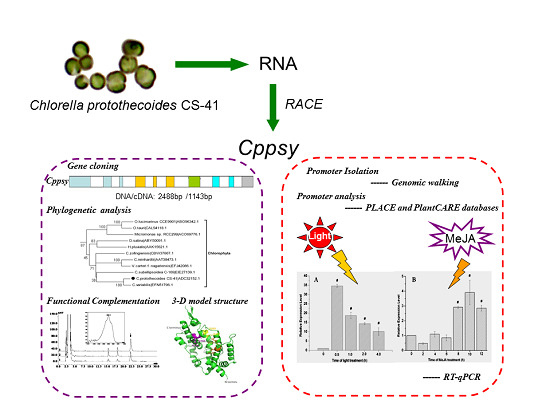
Isolation and Analysis of the Cppsy Gene and Promoter from Chlorella protothecoides CS-41
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cloning and Characterization of the psy Gene from C. protothecoides
| Aim | Primer Sequence 5′-3′ |
|---|---|
| Partial psy fragment | |
| YF | GCCATCTACGTGTGGTGCC |
| YR | CACGCAAGATGTTGGTCAGC |
| 5′-RACE-PCR | |
| YFO1 | GACTTGTCCACGCCCATCAC |
| YRI1 | GGGGAAGCGGGAGATGGTGT |
| 3′-RACE-PCR | |
| YFO2 | GATGCTGCCCTCACAGACAC |
| YRI2 | TGGATTTGGTCAAGTCACGC |
| cDNA and DNA | |
| YF1 | ATGAGCACGTTTCTGAGCACAGTG |
| YR1 | TCACATGCGCGCCCTCAG |
| Probe | |
| psy-F | GAAGTGACCAGCGAGTATGCC |
| psy-R | CTAAAGGGTTGGATGTGC |
| psyRTF | GAAGTGACCAGCGAGTATGCC |
| psyRTR | TCTCTAAAGGGTTGGATGTGC |
| Promoter | |
| PSYSP1 | CTGTGCATGCGAAGTCGGAGTGAGA |
| PSYSP2 | CGTCTTGGCATACTCGCTGGTCACTT |
| PSYSP3 | ACTCATGCTGGGGGCTAGGAAAG |
| PSYSP1′ | ATGGCGGGTGGCAGAGTCAATGTA C |
| PSYSP2′ | CCAGACACAATCACCTCGCAGCCCTT |
| PSYSP3′ | CGTTCACTCACCGCTCTCCATCACAA |

2.2. Sequence Alignment and Phylogenetic Reconstruction


2.3. Promoter Isolation and Analysis

2.4. Gene Expression Response to Light and MeJA

3. Experimental Section
3.1. Strains and Culture Conditions
3.2. Genomic DNA and RNA Isolation
3.3. Cloning of Full-Length Cppsy cDNA and Its Corresponding Gene
3.4. Southern Blot Analysis
3.5. Bioinformatics Analysis
3.6. Functional Complementation Experiment in E. coli
| Strains or Plasmids | Characteristics | Source |
|---|---|---|
| E. coli JM109 | Host for expression vectors | MOST-USDA Joint Research Center for Food Safety stock |
| pACCRT-E | pACYC184 containing crtE gene of Erwinia uredovora (Cmr) (metabolite: GGPP) | Gift from Prof. Gerhard. Sandmann (J.W. Goethe University, Frankfurt, Germany) |
| pACCRT-EB | pACYC184 containing crtE and ctrB genes of Erwinia uredovora (Cmr) (metabolite: Phytoene) | Gift from Prof. Gerhard. Sandmann (J.W. Goethe University, Frankfurt, Germany) |
| pUC19 | Expression vector (Ampr) | MOST-USDA Joint Research Center for Food Safety stock |
| pUC-psy | pUC19 vector containing Cppsy gene (Ampr) | This work |
3.7. Promoter Isolation and Analysis
3.8. Gene Expression Response to Light and MeJA
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflict of Interest
References
- Demmig-Adams, B.; Adams, W.W. Antioxidants in photosynthesis and human nutrition. Science 2002, 298, 2149–2153. [Google Scholar] [CrossRef] [PubMed]
- Vishwanathan, R.; Kuchan, M.J.; Sen, S.; Johnson, E.J. Lutein and Preterm Infants with Decreased Concentrations of Brain Carotenoids. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Perrone, S.; Tei, M.; Longini, M.; Santacroce, A.; Turrisi, G.; Proietti, F.; Felici, C.; Picardi, A.; Bazzini, F.; Vasarri, P.; et al. Lipid and protein oxidation in newborn infants after lutein administration. Oxid. Med. Cell. Longev. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Kalariya, N.M.; Ramana, K.V.; Vankuijk, F.J. Focus on molecules: Lutein. Exp. Eye Res. 2012, 102, 107–108. [Google Scholar] [CrossRef] [PubMed]
- Widomska, J.; Subczynski, W.K. Why has Nature Chosen Lutein and Zeaxanthin to Protect the Retina? J. Clin. Exp. Ophthalmol. 2014, 5, 326. [Google Scholar] [CrossRef] [PubMed]
- Mares-Perlman, J.A.; Millen, A.E.; Ficek, T.L.; Hankinson, S.E. The body of evidence to support a protective role for lutein and zeaxanthin in delaying chronic disease: Overview. J. Nutr. 2002, 132, 518S–524S. [Google Scholar] [PubMed]
- Kim, H.W.; Chew, B.P.; Wong, T.S.; Park, J.S.; Weng, B.B.; Byrne, K.M.; Hayek, M.G.; Reinhart, G.A. Dietary lutein stimulates immune response in the canine. Vet. Immunol. Immunopathol. 2000, 74, 315–327. [Google Scholar] [CrossRef]
- Krinsky, N.I.; Landrum, J.T.; Bone, R.A. Biologic mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu. Rev. Nutr. 2003, 23, 171–201. [Google Scholar] [CrossRef] [PubMed]
- Granado, F.; Olmedilla, B.; Blanco, I. Nutritional and clinical relevance of lutein in human health. Annu. Rev. Nutr. 2003, 90, 487–502. [Google Scholar] [CrossRef]
- Maia, M.; Furlani, B.A.; Souza-Lima, A.A.; Martins, D.S.; Navarro, R.M.; Belfort, R.J. Lutein: A new dye for chromovitrectomy. Retina 2014, 34, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.M.; Zhang, X.W.; Chen, F. Heterotrophic production of biomass and lutein by Chlorella protothecoides on various nitrogen sources. Enzyme Microb. Technol. 2000, 27, 312–318. [Google Scholar] [CrossRef]
- Shi, X.M.; Jiang, Y.; Chen, F. High-yield production of lutein by the green microalga Chlorella protothecoides in heterotrophic fed-batch culture. Biotechnol. Prog. 2002, 18, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.M.; Wu, Z.Y.; Chen, F. Kinetic modeling of lutein production by heterotrophic Chlorella at various pH and temperatures. Mol. Nutr. Food Res. 2006, 50, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Jiang, J.G.; Wang, F. Molecular phylogenies and evolution of crt genes in algae. Crit. Rev. Biotechnol. 2007, 27, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Salvini, M.; Bernini, A.; Fambrini, M.; Pugliesi, C. cDNA cloning and expression of the phytoene synthase gene in sunflower. J. Plant Physiol. 2005, 162, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vallabhaneni, R.; Yu, J.; Rocheford, T.; Wurtzel, E.T. The maize phytoene synthase gene family: Overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008, 147, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Bohne, F.; Linden, H. Regulation of carotenoid biosynthesis genes in response to light in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 2002, 1579, 26–34. [Google Scholar] [CrossRef]
- Lao, Y.M.; Xiao, L.; Ye, Z.W.; Jiang, J.G.; Zhou, S.S. In silico analysis of phytoene synthase and its promoter reveals hints for regulation mechanisms of carotenogenesis in Duanliella bardawil. Bioinformatics 2011, 27, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, J.; Linden, H. Regulation of two carotenoid biosynthesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis. Plant Physiol. 2001, 125, 810–817. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.S.; Kobayashi, M.C.; Niyogi, K.K. White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics 2004, 168, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, J.; Linden, H. Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: Regulation by photosynthetic redox control. Plant Mol. Biol. 2003, 52, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Couso, I.; Vila, M.; Rodriguez, H.; Vargas, M.A.; Leon, R. Overexpression of an exogenous phytoene synthase gene in the unicellular alga Chlamydomonas reinhardtii leads to an increase in the content of carotenoids. Biotechnol. Prog. 2011, 27, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Cordero, B.F.; Couso, I.; Leon, R.; Rodriguez, H.; Vargas, M.A. Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl. Microbiol. Biotechnol. 2011, 91, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Gan, Z.B.; Cui, Y.; Shi, C.L.; Shi, X.M. Structure and function characterization of the phytoene desaturase related to the lutein biosynthesis in Chlorella protothecoides CS-41. Mol. Biol. Rep. 2013, 40, 3351–3361. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Gan, Z.B.; Cui, Y.; Shi, C.L.; Shi, X.M. Cloning and characterization of the zeta-carotene desaturase gene from Chlorella protothecoides CS-41. J. Biomed. Biotechnol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, C.E.; Matthews, P.D.; Li, F.; Wurtzel, E.T. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 2004, 135, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vallabhaneni, R.; Wurtzel, E. PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 2008, 146, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Wust, F.; Bar, C.; al-Babili, S.; Beyer, P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008, 147, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.; Haven, J.; Qiu, W.G.; Polle, J.E. An update on carotenoid biosynthesis in algae: Phylogenetic evidence for the existence of two classes of phytoene synthase. Planta 2009, 229, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987, 15, 6643–6653. [Google Scholar] [CrossRef] [PubMed]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.M.; Chen, F.; Yuan, J.-P.; Chen, H. Heterotrophic production of lutein by selected Chlorella strains. J. Appl. Phycol. 1997, 9, 445–450. [Google Scholar] [CrossRef]
- Stewart, C.J.; Via, L.E. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 1993, 14, 748–750. [Google Scholar] [PubMed]
- Yeku, O.; Frohman, M.A. Rapid amplification of cDNA ends (RACE). In RNA; Nielsen, H., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 107–122. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Breitenbach, J.; Zhu, C.; Sandmann, G. Bleaching herbicide norflurazon inhibits phytoene desaturase by competition with the cofactors. J. Agric. Food Chem. 2001, 49, 5270–5272. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.C.; Liu, J.; Li, Y.T.; Chen, F. Isolation and characterization of the phytoene desaturase gene as a potential selective marker for genetic engineering of the astaxanthin-producing green alga Chlorella zofingiensis (Chlorophyta). J. Phycol. 2008, 44, 684–690. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Cui, Y.; Gan, Z.; Shi, C.; Shi, X. Isolation and Analysis of the Cppsy Gene and Promoter from Chlorella protothecoides CS-41. Mar. Drugs 2015, 13, 6620-6635. https://doi.org/10.3390/md13116620
Li M, Cui Y, Gan Z, Shi C, Shi X. Isolation and Analysis of the Cppsy Gene and Promoter from Chlorella protothecoides CS-41. Marine Drugs. 2015; 13(11):6620-6635. https://doi.org/10.3390/md13116620
Chicago/Turabian StyleLi, Meiya, Yan Cui, Zhibing Gan, Chunlei Shi, and Xianming Shi. 2015. "Isolation and Analysis of the Cppsy Gene and Promoter from Chlorella protothecoides CS-41" Marine Drugs 13, no. 11: 6620-6635. https://doi.org/10.3390/md13116620
APA StyleLi, M., Cui, Y., Gan, Z., Shi, C., & Shi, X. (2015). Isolation and Analysis of the Cppsy Gene and Promoter from Chlorella protothecoides CS-41. Marine Drugs, 13(11), 6620-6635. https://doi.org/10.3390/md13116620