Establishing the Secondary Metabolite Profile of the Marine Fungus: Tolypocladium geodes sp. MF458 and Subsequent Optimisation of Bioactive Secondary Metabolite Production
Abstract
:1. Introduction
2. Results
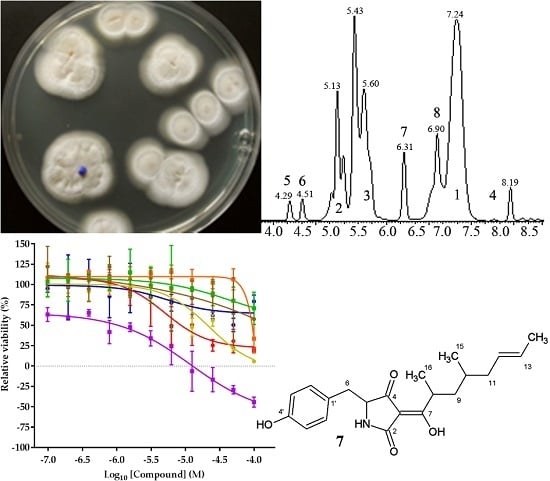
2.1. Classification of Strain MF458
2.2. Isolation and Identification of 1–6
2.3. Structure Elucidation of 7 and 8
2.4. Biological Activities of 1–8
2.5. Secondary Metabolite Profile of MF458 Depending on Fermentation Conditions
2.6. Optimisation of Production for Scaling-Up the Production of the Malettinins
3. Discussion
3.1. Phylogenetic Position
3.2. Secondary Metabolite Profile
3.3. Biological Activity Profile
3.4. Fermentation Optimization
4. Methods
4.1. Isolation and Taxonomy of MF458
4.2. Fermentation and Crude Extract Production
4.3. Anti-Tumour Assay-Guidance for Purification and Pure Compound Evaluation
4.4. Purification and Identification of Compounds 1–8
4.5. Assessment of Compound Cytotoxicity
4.6. General
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A

| Properties | Tolypocladenol C |
|---|---|
| Appearance | Pale yellow semi-solid/gum |
| UV (MeOH) λmax (log ε) | 219 (3.97), 241sh (3.89), 280 (3.95) nm |
| ESI+-MS (m/z) | 358 (MH+) |
| ESI−-MS (m/z) | 356 ([M − H]−) |
| Molecular formula | C21H27NO4 |
| HRESI+-MS (m/z) | |
| Found for MH+ | 358.2016 |
| Calculated | 358.2013 |
| [α]D20 | −135.2° (c 0.016, MeOH) |
| IR µmax | 3250, 2963, 2920, 1654, 1603, 1516, 1451, 1378, 1336, 1269, 1238, 1172, 1105, 1018, 968, 952, 816 cm−1 |
| No. | δ 1H (J [Hz]) | δ 13C |
|---|---|---|
| 1 | ||
| 2 | 176.7 | |
| 3 | 103.0 | |
| 4 | 198.2 | |
| 5 | 3.95 dd (6.4, 3.9) | 63.0 |
| 6 | 2.98 dd (13.9, 4.2) 2.82 dd (13.9, 6.0) | 37.9 |
| 7 | 197.3 | |
| 8 | 3.79 m | 36.5 |
| 9 | 1.72 m 1.04 dd (8.8, 4.4) | 41.6 |
| 10 | 1.29 m | 32.5 |
| 11 | 1.95 m 1.77 m | 41.5 |
| 12 | 5.38 m | 130.9 |
| 13 | 5.38 m | 127.0 |
| 14 | 1.63 m | 18.1 |
| 15 | 0.80 d (6.5) | 20.0 |
| 16 | 1.01 d (6.8) | 18.8 |
| 1′ | 128.1 | |
| 2′ | 6.99 d (8.4) | 131.8 |
| 3′ | 6.65 d (8.4) | 116.0 |
| 4′ | 157.2 | |
| 5′ | 6.65 d (8.4) | 116.0 |
| 6′ | 6.99 d (8.4) | 131.8 |
| Cell Line | Origin | Value | Cyclosporin A | Efrapeptin D | Pyridoxatin | Malettinin B | Malettinin E | Tolypocladenol C | Tolypocladenols A1/A2 |
|---|---|---|---|---|---|---|---|---|---|
| UO-31 | Renal carcinoma | GI50 | 18 ± 0.8 | 6.5 ± 7.5 | 4.4 ± 0.5 | 34.5 ± 21.1 | 33 ± 3.6 | 49.1 ± 21.8 | nd |
| TGI | 99 | 59 ± 56.6 | 9.6 ± 2.3 | 59 | 638.6 ± 143.5 | 214.4 ± 114.6 | nd | ||
| LC50 | 310 | 240 | 26 | 98 | # | 440 | nd | ||
| 786-O | Renal adenocarinoma | GI50 | 46 | 1.6 ± 1 | 3.9 ± 1.6 | 20.4 ± 5.3 | 13.1 ± 2.3 | 75 | 19.6 ± 19 |
| TGI | # | 20 ± 21.1 | 10.1 ± 5.8 | 25 | 29.2 ± 10.1 | # | 149 ± 114.6 | ||
| LC50 | # | 325 ± 445.5 | 21.5 ± 14.7 | # | 142.5 ± 106.1 | # | # | ||
| TK10 | Renal carcinoma | GI50 | 17 ± 3.4 | 3.7 ± 4.4 | 12.2 ± 7 | 19.8 ± 5.9 | 6.6 ± 1.3 | 97.5 ± 55 | nd |
| TGI | 49 | 800 | 74 ± 28.2 | # | # | 104.5 ± 58.5 | nd | ||
| LC50 | # | # | 250 ± 134.5 | # | # | 110.5 ± 63.1 | nd | ||
| SN12C | Renal carcinoma | GI50 | # | 1.9 ± 0.4 | 6.3 ± 0.8 | 20.5 ± 10.6 | 16 ± 7.1 | # | nd |
| TGI | # | 10.2 ± 4 | 17.3 ± 15.1 | 57 | 45 | # | nd | ||
| LC50 | # | # | # | # | 120 | # | nd | ||
| UACC-62 | Malignant melanoma | GI50 | # | # | 9.2 ± 1.1 | 38 ± 2.8 | 10.5 ± 0.7 | 58.5 ± 13.4 | nd |
| TGI | # | 0.4 ± 0.3 | 22 ± 7.1 | 450 | # | 78 ± 9.9 | nd | ||
| LC50 | # | # | 51 | # | # | 92 ± 7.2 | nd | ||
| SK-MEL-28 | Malignant melanoma | GI50 | # | 1.3 ± 1.4 | 11 ± 11.3 | # | 61.5 ± 30 | 7.8 | 46 |
| TGI | 2.9 | 11 | 62.7 ± 58.9 | # | 760 | # | 51 | ||
| LC50 | # | # | 42 | # | # | # | 52 | ||
| M14 | Amelanotic melanoma | GI50 | 12.5 ± 1 | 2.1 ± 1.1 | 2.2 ± 1.6 | 13.7 ± 2.5 | 9.6 ± 4.6 | # | nd |
| TGI | 67 ± 24.5 | 39 | 4.1 ± 2.4 | 102 ± 25.5 | 36 ± 10.1 | # | nd | ||
| LC50 | # | 440 | 25 ± 5.7 | 370 | 244.5 ± 259.4 | # | nd | ||
| U251 | Glioblastoma | GI50 | 40 ± 24 | # | 4.5 ± 0.8 | 18.5 ± 0.7 | 13.5 ± 2.1 | 56.5 ± 6.4 | nd |
| TGI | # | 5.6 | 8.2 ± 4 | # | 48.5 ± 30.4 | 64 ± 14.1 | nd | ||
| LC50 | # | # | 24.5 ± 10.6 | # | # | 69 ± 19.8 | nd | ||
| SF-539 | Gliosarcoma | GI50 | 12.5 ± 1.3 | 1.2 ± 0.7 | 5 ± 1.2 | 14.5 ± 2.1 | 9.4 ± 2.6 | # | nd |
| TGI | 17.5 ± 0.7 | 61.6 ± 116.8 | 12.3 ± 7.5 | 47 ± 18.4 | 27.2 ± 10.9 | # | nd | ||
| LC50 | 26 | 101.3 ± 155.1 | 40 ± 1.4 | # | 250 | # | nd | ||
| RPMI 8226 | Multiple myeloma | GI50 | 7.5 ± 2.8 | # | 2.9 ± 0.4 | 49.5 ± 0.7 | 27.5 ± 3.5 | # | 19.5 ± 7.8 |
| TGI | 12 ± 2.8 | # | 5.2 ± 0.6 | 53 ± 2.8 | 31 ± 7.1 | # | 38 ± 29.7 | ||
| LC50 | 34 | 0.7 ± 0.3 | 8.5 ± 0.8 | 55.5 ± 4.9 | # | # | # | ||
| HL-60 | Procyelo-cytic leukemia | GI50 | 14.3 ± 2.6 | 0.9 ± 1.3 | 3.1 ± 2.4 | 29 ± 5.2 | 11.4 ± 4 | # | nd |
| TGI | 13.5 ± 0.7 | 2.1 ± 2.3 | 3.3 ± 2.5 | 37.3 ± 4.5 | 14.7 ± 3.7 | # | nd | ||
| LC50 | 14.5 ± 2.1 | 7.5 ± 3.1 | 3.7 ± 2.6 | 48 ± 6.6 | 20.4 ± 6.1 | # | nd | ||
| OVCAR-5 | Ovarian carcinoma | GI50 | # | 0.2 ± 0.1 | 4.5 ± 2.9 | 50 ± 1.4 | 20.5 ± 18.1 | # | 41.5 ± 16.3 |
| TGI | # | 5.8 ± 2.9 | 31 ± 9.9 | 60.5 ± 13.4 | 54.7 ± 2.1 | # | 68 | ||
| LC50 | # | 66.5 ± 44.8 | 101.5 ± 26.2 | 69.5 ± 24.7 | 64 ± 11.3 | # | 81 | ||
| OVCAR-3 | Ovarian carcinoma | GI50 | # | 5.8 ± 9.7 | 9.6 ± 20.6 | 22 ± 12.5 | 16.4 ± 9.6 | 69 ± 34.2 | 57 ± 14.1 |
| TGI | # | 273.3 ± 240.9 | 3.6 ± 2.3 | 25.5 ± 20.5 | 16.1 ± 6.9 | 84.7 ± 47.9 | 111 ± 41 | ||
| LC50 | 43 | 57.6 ± 97.3 | 28.3 ± 12.1 | # | # | 89 ± 52.8 | 175 ± 77.8 | ||
| IGR-OV1 | Ovarian adeno-carcinoma | GI50 | 5.3 ± 4.8 | 0.3 ± 0.1 | 3.5 ± 2.4 | 32.7 ± 26.5 | 19 ± 18.8 | # | # |
| TGI | 145 ± 7.1 | 30.3 ± 33.6 | 11 ± 2.6 | 51 | 63 ± 23.1 | # | # | ||
| LC50 | # | 1.7 ± 1.2 | 47.3 ± 17.1 | 53.5 ± 0.7 | 94 ± 74.5 | # | # | ||
| MDA-MB-468 | Breast cancer | GI50 | 5.2 ± 1.5 | # | 2.4 ± 2 | 14.3 ± 5.1 | 6.1 ± 0.8 | 72.8 ± 38.9 | nd |
| TGI | 10.5 ± 1.1 | 0.2 | 3.3 ± 1.6 | 18.8 ± 3.2 | 8.2 ± 1.5 | 96.3 ± 50.6 | nd | ||
| LC50 | 29.3 ± 12.1 | 330 | 4.8 ± 1.1 | 27.3 ± 4.6 | 11.3 ± 2.7 | 120.5 ± 79.8 | nd | ||
| MCF-7 | Breast adeno-carcinoma | GI50 | 18.3 ± 4.9 | 0.3 | 2.3 ± 1.3 | 13.5 ± 2.1 | 10.2 ± 3.8 | 70.5 ± 20.5 | 120 |
| TGI | # | 0.8 | 4.8 ± 3.5 | 19.3 ± 6.7 | 11.6 ± 3.4 | 100 ± 28.3 | 140 | ||
| LC50 | # | 2.8 | 8.1 ± 6.9 | 38.3 ± 28.4 | 15.5 ± 2.1 | 150 | 150 | ||
| HCT-15 | Colon adenocarinoma | GI50 | # | 11 ± 22 | 2.6 | 28 ± 1.4 | 9.4 ± 10.8 | 42 ± 1.4 | 46 ± 1.4 |
| TGI | # | 16.4 ± 28.3 | 5.5 | 32 ± 2.8 | 25.5 ± 0.7 | 49.5 ± 0.7 | 49.5 ± 0.7 | ||
| LC50 | # | 17 ± 29.4 | 12 | 37 ± 5.7 | 31 ± 7.1 | 52 | 51.5 ± 0.7 | ||
| DU145 | Prostate carinoma | GI50 | 11.3 ± 2.4 | 0.6 ± 0.8 | 4.1 | 34.4 ± 8.4 | 16.9 ± 9.4 | 87.3 ± 33.2 | nd |
| TGI | 74 | 267.7 ± 229.9 | 18 | 62 ± 5.7 | 41.4 ± 24.6 | 123.4 ± 61.8 | nd | ||
| LC50 | # | 455 ± 91.9 | 46 | 86 | 94 ± 6.9 | 156.6 ± 89.9 | nd |
| Cell Line | Seeding Cell Density | CELL LINE | Initial Cell Density |
|---|---|---|---|
| UO-31 | 750 cells per well | RPMI8226 | 1200 cells per well |
| 786-O | 750 cells per well | HL-60 | 2000 cells per well |
| TK10 | 1200 cells per well | OVCAR-5 | 500 cells per well |
| SN12C | 1200 cells per well | OVCAR-3 | 2000 cells per well |
| UACC-62 | 1200 cells per well | IGR-OV1 | 500 cells per well |
| SK-MEL-28 | 1000 cells per well | MDA-MB-468 | 1200 cells per well |
| M14 | 2000 cells per well | MCF-7 | 2000 cells per well |
| U251 | 750 cells per well | HCT-15 | 400 cells per well |
| SF-539 | 1000 cells per well | DU145 | 1200 cells per well |
References
- Hickford, S.J.; Blunt, J.W.; Munro, M.H. Antitumour polyether macrolides: Four new halichondrins from the new zealand deep-water marine sponge lissodendoryx sp. Bioorg. Med. Chem. 2009, 17, 2199–2203. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2008, 25, 35–94. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F.; Stohr, R. Sponge-associated bacteria: General overview and special aspects of bacteria associated with halichondria panicea. Prog. Mol. Subcell. Biol. 2003, 37, 35–57. [Google Scholar] [PubMed]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2004, 21, 519–538. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Ishibashi, M. Bioactive metabolites of symbiotic marine microorganisms. Chem. Rev. 1993, 93, 1753–1769. [Google Scholar] [CrossRef]
- Bhakuni, D.S.; Rawat, D.S. Bioactive Marine Natural Products; Springer: New York, NY, USA, 2005; p. 382. [Google Scholar]
- Proksch, P.; Edrada, R.A.; Ebel, R. Drugs from the seas–current status and microbiological implications. Appl. Microbiol. Biotechnol. 2002, 59, 125–134. [Google Scholar] [PubMed]
- Gomes, N.G.; Dasari, R.; Chandra, S.; Kiss, R.; Kornienko, A. Marine invertebrate metabolites with anticancer activities: Solutions to the “supply problem”. Mar. Drugs 2016, 14, 98. [Google Scholar] [CrossRef] [PubMed]
- Schaufelberger, D.E.; Koleck, M.P.; Beutler, J.A.; Vatakis, A.M.; Alvarado, A.B.; Andrews, P.; Marzo, L.V.; Muschik, G.M.; Roach, J.; Ross, J.T.; et al. The large-scale isolation of bryostatin 1 from bugula neritina following current good manufacturing practices. J. Nat. Prod. 1991, 54, 1265–1270. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.; Cragg, G. Natural products in medicinal chemistry. Bioorg. Med. Chem. 2009, 17, 2120. [Google Scholar] [CrossRef] [PubMed]
- Bahn, Y.S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. Microbiol. 2007, 5, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, C.; D’Souza-Ticlo, D.; Verma, A.K. Treatment of colored effluents with lignin-degrading enzymes: An emerging role of marine-derived fungi. Crit. Rev. Microbiol. 2008, 34, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Sprote, P.; Al-Abdallah, Q.; Gehrke, A.; Plattner, H.; Tuncher, A. Regulation of penicillin biosynthesis in filamentous fungi. Adv. Biochem. Eng. Biotechnol. 2004, 88, 45–90. [Google Scholar] [PubMed]
- Imhoff, J.F. Natural products from marine fungi—Still an underrepresented resource. Mar. Drugs 2016, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R.; Rowley, D.C. Halovir, an Antiviral Marine Natural Product, and Derivatives Thereof. U.S. Patent 6,458,766 B1, 1 October 2002. [Google Scholar]
- Holler, U.; Konig, G.M.; Wright, A.D. Three new metabolites from marine-derived fungi of the genera coniothyrium and microsphaeropsis. J. Nat. Prod. 1999, 62, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Ali, M.S.; Hussain, S.; Jabbar, A.; Ashraf, M.; Lee, Y.S. Marine natural products of fungal origin. Nat. Prod. Rep. 2007, 24, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.G.; Lefranc, F.; Kijjoa, A.; Kiss, R. Can some marine-derived fungal metabolites become actual anticancer agents? Mar. Drugs 2015, 13, 3950–3991. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, B.; Silber, J.; Prashar, A.; Landskron, J.; Weber, J.; Rehermann, S.; Muller, F.J.; Smith, S.; Wrigley, S.; Tasken, K.; et al. A phenotypic screening approach to identify anticancer compounds derived from marine fungi. Assay Drug Dev. Technol. 2014, 12, 162–175. [Google Scholar] [PubMed]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Hofs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Lee, K.H.; Sanchez, J.F.; Keller, N.P.; Wang, C.C. Unlocking fungal cryptic natural products. Nat. Prod. Commun. 2009, 4, 1505–1510. [Google Scholar] [PubMed]
- Survase, S.A.; Kagliwal, L.D.; Annapure, U.S.; Singhal, R.S. Cyclosporin a—A review on fermentative production, downstream processing and pharmacological applications. Biotechnol. Adv. 2011, 29, 418–435. [Google Scholar] [CrossRef] [PubMed]
- Bandani, A.R.; Khambay, B.P.S.; Faull, J.L.; Newton, R.; Deadman, M.; Butt, T.M. Production of efrapeptins by tolypocladium species and evaluation of their insecticidal and antimicrobial properties. Mycol. Res. 2000, 104, 537–544. [Google Scholar] [CrossRef]
- Kessler, H.; Loosli, H.R.; Oschkinat, H. Peptide conformations. 30. Assignment of the h-1-nmr, c-13-nmr, and n-15-nmr spectra of cyclosporin-a in cdcl3 and c6d6 by a combination of homonuclear and heteronuclear two-dimensional techniques. Helv. Chim. Acta 1985, 68, 661–681. [Google Scholar] [CrossRef]
- Gupta, S.; Krasnoff, S.B.; Roberts, D.W.; Renwick, J.A.A.; Brinen, L.S.; Clardy, J. Structures of the efrapeptins: Potent inhibitors of mitochondrial atpase from the fungus tolypocladium-niveum. J. Am. Chem. Soc. 1991, 113, 707–709. [Google Scholar] [CrossRef]
- Teshima, Y.; Shinya, K.; Shimazu, A.; Furihata, K.; Chul, H.S.; Furihata, K.; Hayakawa, Y.; Nagai, K.; Seto, H. Isolation and structural elucidation of pyridoxatin, a free-radical scavenger of microbial origin. J. Antibiot. 1991, 44, 685–687. [Google Scholar] [CrossRef] [PubMed]
- Jegorov, A.; Matha, V.; Husak, M.; Kratochvil, B.; Stuchlik, J.; Sedmera, P.; Havlicek, V. Iron uptake system of some members of the genus tolypocladium–crystal-structure of the ligand and its iron(iii) complex. J. Chem. Soc. Dalton 1993, 28, 1287–1293. [Google Scholar] [CrossRef]
- Angawi, R.F.; Swenson, D.C.; Gloer, J.B.; Wicklow, D.T. Malettinins b-d: New polyketide metabolites from an unidentified fungal colonist of hypoxylon stromata (nrrl 29110). J. Nat. Prod. 2005, 68, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Ohlendorf, B.; Labes, A.; Wenzel-Storjohann, A.; Näther, C.; Imhoff, J.F. Malettinin e, an antibacterial and antifungal tropolone produced by a marine cladosporium strain. Front. Mar. Sci. 2014, 1, 1–6. [Google Scholar] [CrossRef]
- Turner, W.B.A.; Aldridge, D.C. Fungal Metabolites ii; Academic Press: New York, NY, USA, 1983. [Google Scholar]
- Li, X.B.; Li, L.; Zhu, R.X.; Li, W.; Chang, W.Q.; Zhang, L.L.; Wang, X.N.; Zhao, Z.T.; Lou, H.X. Tetramic acids and pyridone alkaloids from the endolichenic fungus tolypocladium cylindrosporum. J. Nat. Prod. 2015, 78, 2155–2160. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.; Riese, U.; Li, Z.Z.; Hamburger, M. Novel tetramic acids and pyridone alkaloids, militarinones b, c, and d, from the insect pathogenic fungus paecilomyces militaris. J. Nat. Prod. 2003, 66, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Wangun, H.V.K.; Hertweck, C. Epicoccarines a, b and epipyridone: Tetramic acids and pyridone alkaloids from an epicoccum sp associated with the tree fungus pholiota squarrosa. Org. Biomol. Chem. 2007, 5, 1702–1705. [Google Scholar] [CrossRef] [PubMed]
- Kakule, T.B.; Zhang, S.; Zhan, J.; Schmidt, E.W. Biosynthesis of the tetramic acids sch210971 and sch210972. Org. Lett. 2015, 17, 2295–2297. [Google Scholar] [CrossRef] [PubMed]
- Gledhill, J.R.; Walker, J.E. Inhibitors of the catalytic domain of mitochondrial atp synthase. Biochem. Soc. Trans. 2006, 34, 989–992. [Google Scholar] [CrossRef] [PubMed]
- Angawi, R.F.; Swenson, D.C.; Gloer, J.B.; Wicklow, D.T. Malettinin a: A new antifungal tropolone from an unidentified fungal colonist of hypoxylon stromata (nrrl 29110). Tetrahedron Lett. 2003, 44, 7593–7596. [Google Scholar] [CrossRef]
- Sung, G.H.; Spatafora, J.W.; Zare, R.; Hodge, K.T.; Gams, W. A revision of verticillium sect. Prostrata. Ii. Phylogenetic analyses of ssu and lsu nuclear rdna sequences from anamorphs and teleomorphs of the clavicipitaceae. Nova Hedwigia 2001, 72, 311–328. [Google Scholar]
- Quandt, C.A.; Kepler, R.M.; Gams, W.; Araujo, J.P.; Ban, S.; Evans, H.C.; Hughes, D.; Humber, R.; Hywel-Jones, N.; Li, Z.; et al. Phylogenetic-based nomenclatural proposals for ophiocordycipitaceae (hypocreales) with new combinations in tolypocladium. IMA Fungus 2014, 5, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Gams, W. Cephalosporium-Artige Schimmelpilze (Hyphomycetes); G. Fischer: Stuttgart, Germany, 1971. [Google Scholar]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D.L. A new dawn for the naming of fungi: Impacts of decisions made in melbourne in july 2011 on the future publication and regulation of fungal names. Mycokeys 2011, 1, 7–20. [Google Scholar] [CrossRef]
- Kramer, A.; Labes, A.; Imhoff, J.F. Phylogenetic relationship and secondary metabolite production of marine fungi producing the cyclodepsipeptides scopularide a and b. Mar. Biotechnol. 2016, 18, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Ohlendorf, B.; Erhard, A.; Labes, A.; Imhoff, J.F. Bioactive Metabolites of a Marine Calcarisporium sp. In Proceedings of the Research & Innovation-Inpact into Cancer, Shropshire, UK, 11 March 2014. [Google Scholar]
- Bushley, K.E.; Raja, R.; Jaiswal, P.; Cumbie, J.S.; Nonogaki, M.; Boyd, A.E.; Owensby, C.A.; Knaus, B.J.; Elser, J.; Miller, D.; et al. The genome of tolypocladium inflatum: Evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet. 2013, 9, e1003496. [Google Scholar] [CrossRef] [PubMed]
- Mascarell, L.; Truffa-Bachi, P. New aspects of cyclosporin a mode of action: From gene silencing to gene up-regulation. Mini Rev. Med. Chem. 2003, 3, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Papathanassiu, A.E.; MacDonald, N.J.; Emlet, D.R.; Vu, H.A. Antitumor activity of efrapeptins, alone or in combination with 2-deoxyglucose, in breast cancer in vitro and in vivo. Cell. Stress Chaperon 2011, 16, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Chung, M.C.; Lee, C.H.; Chun, H.K.; Kim, H.M.; Kho, Y.H. Pyridoxatin, an inhibitor of gelatinase a with cytotoxic activity. J. Microbiol. Biotechnol. 1996, 6, 445–450. [Google Scholar]
- Cai, P.; Smith, D.; Cunningham, B.; Brown-Shimer, S.; Katz, B.; Pearce, C.; Venables, D.; Houck, D. 8-methyl-pyridoxatin: A novel n-hydroxy pyridone from fungus os-f61800 that induces erythropoietin in human cells. J. Nat. Prod. 1999, 62, 397–399. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Zhang, M.; Li, Y.; Li, X.; Gao, Y.; Xie, Z.; Lou, H. Lichen endophyte derived pyridoxatin inactivates candida growth by interfering with ergosterol biosynthesis. Biochim. Biophys. Acta 2015, 1850, 1762–1771. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, G.; Gulder, T.A.; Lang, G.; Schmitt, S.; Stohr, R.; Wiese, J.; Nagel, K.; Imhoff, J.F. Large-scale biotechnological production of the antileukemic marine natural product sorbicillactone a. Mar. Drugs 2007, 5, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, G.; Lang, G.; Gulder, T.A.M.; Tsuruta, H.; Muhlbacher, J.; Maksimenka, K.; Steffens, S.; Schaumann, K.; Stohr, R.; Wiese, J.; et al. The first sorbicillinoid alkaloids, the antileukemic sorbicillactones a and b, from a sponge-derived penicillium chrysogenum strain. Tetrahedron 2005, 61, 7252–7265. [Google Scholar] [CrossRef]
- Sears, M.A.; Gerhart, D.J.; Rittschof, D. Antifouling agents from marine spongelissodendoryx isodictyalis carter. J. Chem. Ecol. 1990, 16, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [PubMed]
- Tatusova, T.A.; Madden, T.L. Blast 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999, 174, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.B. Mycology Guidebook; University of Washington Press: Seattle, WA, USA, 1974. [Google Scholar]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive investigation of marine actinobacteria associated with the sponge halichondria panicea. Appl. Environ. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef] [PubMed]
- Wickerham, L.J. Taxonomy of Yeasts; U.S. Department of Agriculture: Washington, DC, USA, 1951.
- Shoemaker, R.H. The nci60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef] [PubMed]






| Condition | Malettinin B Yield in mg/L |
|---|---|
| Standing Erlenmeyer flask | 2.28 |
| Shaking Erlenmeyer flask | 2.43 |
| STR without pH control | 18.2 |
| STR with pH control | 10.3 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kebede, B.; Wrigley, S.K.; Prashar, A.; Rahlff, J.; Wolf, M.; Reinshagen, J.; Gribbon, P.; Imhoff, J.F.; Silber, J.; Labes, A.; et al. Establishing the Secondary Metabolite Profile of the Marine Fungus: Tolypocladium geodes sp. MF458 and Subsequent Optimisation of Bioactive Secondary Metabolite Production. Mar. Drugs 2017, 15, 84. https://doi.org/10.3390/md15040084
Kebede B, Wrigley SK, Prashar A, Rahlff J, Wolf M, Reinshagen J, Gribbon P, Imhoff JF, Silber J, Labes A, et al. Establishing the Secondary Metabolite Profile of the Marine Fungus: Tolypocladium geodes sp. MF458 and Subsequent Optimisation of Bioactive Secondary Metabolite Production. Marine Drugs. 2017; 15(4):84. https://doi.org/10.3390/md15040084
Chicago/Turabian StyleKebede, Bethlehem, Stephen K. Wrigley, Anjali Prashar, Janina Rahlff, Markus Wolf, Jeanette Reinshagen, Philip Gribbon, Johannes F. Imhoff, Johanna Silber, Antje Labes, and et al. 2017. "Establishing the Secondary Metabolite Profile of the Marine Fungus: Tolypocladium geodes sp. MF458 and Subsequent Optimisation of Bioactive Secondary Metabolite Production" Marine Drugs 15, no. 4: 84. https://doi.org/10.3390/md15040084
APA StyleKebede, B., Wrigley, S. K., Prashar, A., Rahlff, J., Wolf, M., Reinshagen, J., Gribbon, P., Imhoff, J. F., Silber, J., Labes, A., & Ellinger, B. (2017). Establishing the Secondary Metabolite Profile of the Marine Fungus: Tolypocladium geodes sp. MF458 and Subsequent Optimisation of Bioactive Secondary Metabolite Production. Marine Drugs, 15(4), 84. https://doi.org/10.3390/md15040084