Anti-Phytopathogenic and Cytotoxic Activities of Crude Extracts and Secondary Metabolites of Marine-Derived Fungi
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation, Identification, and Phylogenetic Analysis of the Marine-Derived Fungi
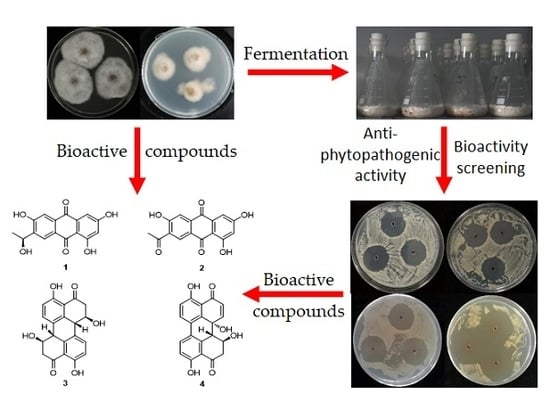
2.2. Screening for Bioactive Marine-Derived Fungal Strains
2.2.1. Antibacterial Activity
2.2.2. Antifungal Activity
2.2.3. Cytotoxicity
2.3. Structure Elucidation of Compounds 1–4
2.4. Bioactivities of Compounds 1–4
2.5. Antibacterial Mechanism of 4
3. Materials and Methods
3.1. Isolation of Marine-Derived Fungi
3.2. DNA Extraction, Polymerase Chain Reaction (PCR) Amplification, and Sequencing
3.3. Phylogenetic Tree Construction
3.4. Fermentation of the Identified Marine-Derived Fungi and Bioactivity Screening
3.5. Isolation and Structure Elucidation of 1–4 from F. equiseti (P18) and Alternaria sp. (P8)
3.6. Biological Assay of the Isolated Compounds
3.6.1. Evaluation of the MIC of the Bioactive Compounds
3.6.2. Cytotoxicity Assay for 1 and 2
3.6.3. Growth Curves Associated with C. michiganensis in the Presence of 4
3.6.4. Nucleotide Leakage
3.6.5. Transmission Electron Microscopy (TEM)
3.6.6. Measurement of Cell Membrane Potential
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sundin, G.W.; Castiblanco, L.F.; Yuan, X.; Zeng, Q.; Yang, C.-H. Bacterial disease management: Challenges, experience, innovation and future prospects. Mol. Plant Pathol. 2016, 17, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.; Ray, A.; Dash, S.; Mishra, A.; Achary, K.G.; Nayak, S.; Singh, S. Fungal disease detection in plants: Traditional assays, novel diagnostic techniques and biosensors. Biosens. Bioelectron. 2017, 87, 708–723. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.G.M.; Lefranc, F.; Kijjoa, A.; Kiss, R. Can some marine-derived fungal metabolites become actual anticancer agents? Mar. Drugs 2015, 13, 3950–3991. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. Natural products from marine fungi-still an underrepresented resource. Mar. Drugs 2016, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koul, M.; Singh, S. Penicillium spp.: Prolific producer for harnessing cytotoxic secondary metabolites. Anti-Cancer Drugs 2017, 28, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Pang, X.; Lin, X.; Tian, Y.; Liang, R.; Wang, J.; Yang, B.; Zhou, X.; Kaliyaperumal, K.; Luo, X.; Tu, Z.; et al. Three new polyketides from the marine sponge-derived fungus Trichoderma sp. SCSIO41004. Nat. Prod. Res. 2017, 32. [Google Scholar] [CrossRef] [PubMed]
- Arnone, A.; Nasini, G.; Merlini, L.; Assante, G. Secondary mold metabolites. Part 16. Stemphyltoxins, new reduced perylenequinone metabolites from Stemphylium botryosum var. Lactucum. J. Chem. Soc. Perkin Trans. 1986, 1, 525–530. [Google Scholar] [CrossRef]
- Okuno, T.; Natsume, I.; Sawai, K.; Sawamura, K.; Furusaki, A.; Matsumoto, T. Structure of antifungal and phytotoxic pigments produced by Alternaria species. Tetrahedron Lett. 1983, 24, 5653–5656. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, Q.; Gao, Y.-Q.; Tang, J.-J.; Zhang, A.-L.; Gao, J.-M. Secondary metabolites from the endophytic Botryosphaeria dothidea of Melia azedarach and their antifungal, antibacterial, antioxidant, and cytotoxic activities. J. Agric. Food Chem. 2014, 62, 3584–3590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, H.-X.; Chen, Y.-C.; Sun, Z.-H.; Li, H.-H.; Li, S.-N.; Yan, M.-L.; Zhang, W.-M. Two new metabolites from the endophytic fungus Alternaria sp. A744 derived from Morinda officinalis. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Bai, J.; Zhong, K.; Huang, Y.; Gao, H. A dual antibacterial mechanism involved in membrane disruption and DNA binding of 2R,3R-dihydromyricetin from pine needles of Cedrus deodara against Staphylococcus aureus. Food Chem. 2017, 218, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Whiteaker, K.L.; Gopalakrishnan, S.M.; Groebe, D.; Shieh, C.-C.; Warrior, U.; Burns, D.J.; Coghlan, M.J.; Scott, V.E.; Gopalakrishnan, M. Validation of FLIPR membrane potential dye for high throughput screening of potassium channel modulators. J. Biomol. Screen. 2001, 6, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Santos, B.S.; Silva, L.C.N.; Silva, T.D.; Rodrigues, J.F.S.; Grisotto, M.A.G.; Santos, C.M.T.; Napoleao, T.H.; Silv, M.V.; Paiva, P.M.G. Application of omics technologies for evaluation of antibacterial mechanisms of action of plant-derived products. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-Y.; Yang, K.-L.; Li, J.; Wang, C.-Y.; Shao, C.-L. Phylogenetic diversity and antibacterial activity of culturable fungi derived from the zoanthid Palythoa haddoni in the South China Sea. Mar. Biotechnol. 2015, 17, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A. Developmental peculiarities and seed-borne endophytes in quinoa: Omnipresent, robust bacilli contribute to plant fitness. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-N.; Shao, C.-L.; Zheng, C.-J.; Chen, Y.-Y.; Wang, C.-Y. Diversity and antibacterial activities of fungi derived from the gorgonian Echinogorgia rebekka from the South China Sea. Mar. Drugs 2011, 9, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Yangui, I.; Boutiti, M.Z.; Boussaid, M.; Messaoud, C. Essential oils of Myrtaceae Species growing wild in Tunisia: Chemical variability and antifungal activity against Biscogniauxia mediterranea, the causative agent of charcoal canker. Chem. Biodivers. 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Di, T.-M.; Yang, S.-L.; Du, F.-Y.; Zhao, L.; Xia, T.; Zhang, X.-F. Cytotoxic and hypoglycemic activity of triterpenoid saponins from Camellia oleifera Abel. seed pomac. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liao, Y.; Tang, C.; Huang, X.; Luo, Z.; Chen, J.; Cai, P. Cytotoxic and antibacterial compounds from the coral-derived fungus Aspergillus tritici SP2-8-1. Mar. Drugs 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiong, P.; Zheng, W.; Zhu, X.; She, Z.; Ding, W.; Li, C. Identification and antifungal activity of compounds from the mangrove endophytic fungus Aspergillus clavatus R7. Mar. Drugs 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Hao, T.; Li, S. In vitro antibacterial activities and mechanism of sugar fatty acid esters against five food-related bacteria. Food Chem. 2015, 187, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, F.; Du, L.; Zhao, T.; Doyle, M.P.; Wang, D.; Zhang, X.; Sun, Z.; Xu, W. Antibacterial and antibiofilm activity of phenyllactic acid against Enterobacter cloacae. Food Control 2018, 84, 442–448. [Google Scholar] [CrossRef]
- Sanchez, E.; Garcia, S.; Heredia, N. Extracts of edible and medicinal plants damage membranes of Vibrio cholerae. Appl. Environ. Microbiol. 2010, 76, 6888–6894. [Google Scholar] [CrossRef] [PubMed]







| Strain | P. syringae | A. avenae | E. carotovora | X. oryzae | R. solanacearum | C. michiganensis |
|---|---|---|---|---|---|---|
| P1 | + | − | + | − | − | − |
| P2 | + | + | + | − | − | − |
| P3 | + | + | ++ | − | − | + |
| P4 | ++ | ++ | ++ | − | − | + |
| P7 | − | − | + | − | − | − |
| P8 | + | + | ++ | + | + | + |
| P9 | ++ | ++ | + | − | − | ++ |
| P10 | − | + | ++ | − | − | + |
| P12 | − | − | − | − | + | − |
| P13 | + | + | ++ | − | − | + |
| P14 | ++ | +++ | ++ | − | − | ++ |
| P15 | − | + | − | + | − | − |
| P16 | + | + | + | − | − | − |
| P17 | − | − | − | − | − | + |
| P18 | +++ | +++ | +++ | − | − | +++ |
| P19 | +++ | +++ | +++ | − | − | ++ |
| P20 | +++ | +++ | +++ | − | − | ++ |
| P23 | − | ++ | − | − | − | − |
| P24 | − | + | + | − | − | + |
| P25 | ++ | +++ | ++ | − | + | ++ |
| P27 | ++ | +++ | ++ | − | − | ++ |
| P28 | + | + | + | − | + | − |
| P29 | +++ | +++ | +++ | − | − | ++ |
| P31 | + | − | + | − | − | + |
| Fungal Strain | P. syringae | A. avenae | E. carotovora | C. michiganensis |
|---|---|---|---|---|
| P8 | + | − | − | − |
| P14 | ++ | − | ++ | + |
| P18 | +++ | +++ | +++ | +++ |
| P19 | +++ | − | ++ | + |
| P20 | +++ | − | − | − |
| P25 | + | − | − | − |
| P27 | + | − | − | − |
| P29 | + | − | − | − |
| Fungal Strain | A. alternata (Fries) Keissler | P. parasitica var. nicotianae Tucker |
|---|---|---|
| P3 | − | + |
| P7 | − | + |
| P8 | − | + |
| P11 | − | + |
| P12 | + | − |
| P14 | − | + |
| P17 | + | − |
| P18 | + | + |
| P19 | − | + |
| P20 | − | + |
| P22 | − | + |
| P24 | − | + |
| P26 | − | + |
| P27 | − | + |
| P29 | + | + |
| P31 | − | + |
| Fungal Strain | Inhibition Rate (%) | ||
|---|---|---|---|
| A549 | HeLa | HepG2 | |
| P3 | 57.25 | 92.29 | 76.60 |
| P9 | 91.87 | 96.84 | 93.11 |
| P10 | 69.19 | 89.70 | 83.19 |
| P12 | 92.70 | 88.88 | 87.29 |
| P15 | 87.49 | 89.77 | 88.62 |
| P19 | 96.06 | 97.43 | 97.77 |
| P20 | 95.94 | 97.60 | 97.82 |
| P21 | 87.02 | 97.08 | 94.53 |
| P23 | 90.23 | 84.12 | 80.28 |
| P24 | 96.07 | 97.56 | 97.86 |
| P25 | 88.98 | − | − |
| P30 | − | 65.29 | − |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, D.-L.; Wang, D.; Tian, X.-Y.; Cao, F.; Li, Y.-Q.; Zhang, C.-S. Anti-Phytopathogenic and Cytotoxic Activities of Crude Extracts and Secondary Metabolites of Marine-Derived Fungi. Mar. Drugs 2018, 16, 36. https://doi.org/10.3390/md16010036
Zhao D-L, Wang D, Tian X-Y, Cao F, Li Y-Q, Zhang C-S. Anti-Phytopathogenic and Cytotoxic Activities of Crude Extracts and Secondary Metabolites of Marine-Derived Fungi. Marine Drugs. 2018; 16(1):36. https://doi.org/10.3390/md16010036
Chicago/Turabian StyleZhao, Dong-Lin, Dan Wang, Xue-Ying Tian, Fei Cao, Yi-Qiang Li, and Cheng-Sheng Zhang. 2018. "Anti-Phytopathogenic and Cytotoxic Activities of Crude Extracts and Secondary Metabolites of Marine-Derived Fungi" Marine Drugs 16, no. 1: 36. https://doi.org/10.3390/md16010036
APA StyleZhao, D. -L., Wang, D., Tian, X. -Y., Cao, F., Li, Y. -Q., & Zhang, C. -S. (2018). Anti-Phytopathogenic and Cytotoxic Activities of Crude Extracts and Secondary Metabolites of Marine-Derived Fungi. Marine Drugs, 16(1), 36. https://doi.org/10.3390/md16010036