Marine Natural Products and Drug Resistance in Latent Tuberculosis
Abstract
:1. Introduction
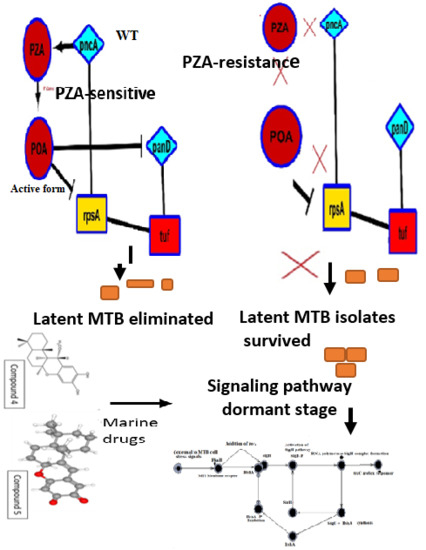
1.1. PZA against Latent TB
1.2. Signaling in Latent TB
1.3. Drugs Effective under Latent Stage
1.4. Marine Natural Products against Latent TB
2. Results
2.1. SigH Regulatory Network
2.2. Paths Identification in the Network
2.3. SigH Regulation and Marine Drugs
3. Discussion
4. Methods
4.1. Literature Search
4.2. Pathway Construction Using Systems Biology Approach
4.3. Validation of SigH Regulatory Pathway
4.4. Synthetic Biology and SigH Activation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2017. Available online: http://apps.who.int/medicinedocs/en/m/abstract/Js23360en/ (accessed on 6 September 2019).
- Ai, J.W.; Ruan, Q.L.; Liu, Q.H.; Zhang, W.H. Updates on the risk factors for latent tuberculosis reactivation and their managements. Emerg. Microbes Infect. 2016, 5, e10. [Google Scholar] [CrossRef] [PubMed]
- Esmail, H.; Barry, C.E.; Wilkinson, R.J. Understanding latent tuberculosis: The key to improved diagnostic and novel treatment strategies. Drug Discov. Today 2012, 17, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Houben, R.M.G.J.; Dodd, P.J. The Global Burden of Latent Tuberculosis Infection: A Re-estimation Using Mathematical Modelling. PLoS Med. 2016, 13, e1002152. [Google Scholar] [CrossRef] [PubMed]
- Fogel, N. Tuberculosis: A disease without boundaries. Tuberculosis 2015, 95, 527–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruchfeld, J.; Correia-Neves, M.; Källenius, G. Tuberculosis and HIV Coinfection. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A. Tuberculosis in HIV/AIDS Patients. Nihon Rinsho 2017, 69, 1433–1437. [Google Scholar]
- Mekonnen, D.; Derbie, A.; Desalegn, E. TB/HIV co-infections and associated factors among patients on directly observed treatment short course in Northeastern Ethiopia: A 4 years retrospective study. BMC Res. Notes 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, A.; Jansson, M.; Sköld, M.; Rottenberg, M.E.; Källenius, G. Tuberculosis and HIV Co-Infection. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Schutz, C.; Meintjes, G.; Almajid, F.; Wilkinson, R.J.; Pozniak, A. Clinical management of tuberculosis and HIV-1 co-infection. Eur. Respir. J. 2010, 36, 1460–1481. [Google Scholar] [CrossRef]
- Tesfaye, B.; Alebel, A.; Gebrie, A.; Zegeye, A.; Tesema, C.; Kassie, B. The twin epidemics: Prevalence of TB/HIV co-infection and its associated factors in Ethiopia; A systematic review and meta-analysis. PLoS ONE 2018, 13, e0203986. [Google Scholar] [CrossRef]
- Anand, R. Identification of Potential Antituberculosis Drugs Through Docking and Virtual Screening. Interdiscip. Sci. Comput. Life Sci. 2018, 10, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Hosen, M.I.; Tanmoy, A.M.; Mahbuba, D.A.; Salma, U.; Nazim, M.; Islam, M.T.; Akhteruzzaman, S. Application of a subtractive genomics approach for in silico identification and characterization of novel drug targets in Mycobacterium tuberculosis F11. Interdiscip. Sci. Comput. Life Sci. 2014, 6, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, D.R.; Shanmughavel, P. In Silico Drug Design of Thiolactomycin Derivatives Against Mtb-KasA Enzyme to Inhibit Multidrug Resistance of Mycobacterium tuberculosis. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.C.; Ma, J.H.; Guthrie, J.L.; Blair, J.; Chedore, P.; Jamieson, F.B. Gene sequencing for routine verification of pyrazinamide resistance in Mycobacterium tuberculosis: A role for pncA but not rpsA. J. Clin. Microbiol. 2012, 50, 3726–3728. [Google Scholar] [CrossRef] [PubMed]
- Allana, S.; Shashkina, E.; Mathema, B.; Bablishvili, N.; Tukvadze, N.; Shah, N.S.; Kempker, R.R.; Blumberg, H.M.; Moodley, P.; Mlisana, K.; et al. pncA Gene Mutations Associated with Pyrazinamide Resistance in Drug-Resistant Tuberculosis, South Africa and Georgia. Emerg. Infect. Dis. 2017, 23, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Aono, A.; Chikamatsu, K.; Yamada, H.; Kato, T.; Mitarai, S. Association between pncA gene mutations, pyrazinamidase activity, and pyrazinamide susceptibility testing in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 4928–4930. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Malik, S.I.; Ali, S.; Sheed Khan, A.; Nadeem, T.; Zeb, M.T.; Masood, N.; Afzal, M.T. Prevalence of Pyrazinamide Resistance in Khyber Pakhtunkhwa, Pakistan. Microb. Drug Resist. 2018. [Google Scholar] [CrossRef] [PubMed]
- Black, G.F.; Thiel, B.A.; Ota, M.O.; Parida, S.K.; Adegbola, R.; Boom, W.H.; Dockrell, H.M.; Franken, K.L.M.C.; Friggen, A.H.; Hill, P.C.; et al. Immunogenicity of novel DosR regulon-encoded candidate antigens of Mycobacterium tuberculosis in three high-burden populations in Africa. Clin. Vaccine Immunol. CVI 2009, 16, 1203–1212. [Google Scholar] [CrossRef]
- Hatzios, S.K.; Baer, C.E.; Rustad, T.R.; Siegrist, M.S.; Pang, J.M.; Ortega, C.; Alber, T.; Grundner, C.; Sherman, D.R.; Bertozzi, C.R. Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc. Natl. Acad. Sci. USA 2013, 110, E5069–E5077. [Google Scholar] [CrossRef]
- Honaker, R.; Dhiman, R.; Narayanasamy, P.; Crick, D.; Voskuil, M. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J. Bacteriol. 2010, 192, 6447–6455. [Google Scholar] [CrossRef]
- Sherman, D.; Voskuil, M.; Schnappinger, D.; Liao, R.; Harrell, M.; Schoolnik, G.; Park, H.; Guinn, K.; Voskuil, M.; Tompa, M.; et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 2001, 98, 7534–7539. [Google Scholar] [CrossRef] [PubMed]
- Park, S.T.; Kang, C.M.; Husson, R.N. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13105–13110. [Google Scholar] [CrossRef] [PubMed]
- Busche, T.; Šilar, R.; Pičmanová, M.; Pátek, M.; Kalinowski, J. Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network in Corynebacterium glutamicum. BMC Genom. 2012, 13, 445. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Farhana, A.; Guidry, L.; Saini, V.; Hondalus, M.; Steyn, A.J. Redox homeostasis in mycobacteria: The key to tuberculosis control? Expert Rev. Mol. Med. 2011, 13, e39. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, A.; Singh, N.; Bhat, S.A.; Gupta, P.; Kumar, A. Redox biology of tuberculosis pathogenesis. Adv. Microb. Physiol. 2012, 60, 263–324. [Google Scholar] [PubMed]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Bashyam, M.; Hasnain, S. The extracytoplasmic function sigma factors: Role in bacterial pathogenesis. Infect. Genet. Evol. 2004, 4, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, N.D.; Wu, Q.L.; Kong, D.; Puyang, X.; Garg, S.; Husson, R.N. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 1999, 181, 4266–4274. [Google Scholar]
- Song, T.; Dove, S.L.; Lee, K.H.; Husson, R.N. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 2003, 50, 949–959. [Google Scholar] [CrossRef]
- Raman, S.; Song, T.; Puyang, X.; Bardarov, S.; Jacobs, J.; Husson, R.N. The alternative sigma factor sigh regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 2001, 183, 6119–6125. [Google Scholar] [CrossRef]
- Graham, J.E.; Clark-Curtiss, J.E. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 1999, 96, 11554–11559. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, D.; Schroeder, B.G.; Tyagi, S.; Yoshimatsu, T.; Scott, C.; Ko, C.; Carpenter, L.; Mehrotra, J.; Manabe, Y.C.; Fleischmann, R.D.; et al. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 2002, 99, 8330–8335. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, D.A. The action of antituberculosis drugs in short-course chemotherapy. Tubercle 1985, 66, 219–225. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Y.; Bi, J.; Cai, Q.; Liao, X.; Li, W.; Guo, C.; Zhang, Q.; Lin, T.; Zhao, Y.; et al. Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide. Mol. Microbiol. 2015, 95, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Yadon, A.N.; Maharaj, K.; Adamson, J.H.; Lai, Y.-P.; Sacchettini, J.C.; Ioerger, T.R.; Rubin, E.J.; Pym, A.S. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Haagsma, A.C.; Pham, H.; Maaskant, J.J.; Mol, S.; Lill, H.; Bald, D. Pyrazinoic Acid Decreases the Proton Motive Force, Respiratory ATP Synthesis Activity, and Cellular ATP Levels. Antimicrob. Agents Chemother. 2011, 55, 5354–5357. [Google Scholar] [CrossRef] [Green Version]
- Ying, Z.; Wade, M.M.; Scorpio, A.; Zhang, H.; Sun, Z. Mode of action of pyrazinamide: Disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J. Antimicrob. Chemother. 2003, 52, 790–795. [Google Scholar]
- Khan, M.T.; Malik, S.I. Structural dynamics behind variants in pyrazinamidase and pyrazinamide resistance. J. Biomol. Struct. Dyn. 2019, 1–15. [Google Scholar] [CrossRef]
- Khan, M. Pyrazinamide resistance and mutations L19R, R140H, and E144K in Pyrazinamidase of Mycobacterium tuberculosis. J. Cell. Biochem. 2019, 120, 7154–7166. [Google Scholar] [CrossRef]
- Khan, M.T.; Khan, A.; Rehman, A.U.; Wang, Y.; Akhtar, K.; Malik, S.I.; Wei, D.Q. Structural and free energy landscape of novel mutations in ribosomal protein S1 (rpsA) associated with pyrazinamide resistance. Sci. Rep. 2019, 9, 7482. [Google Scholar] [CrossRef]
- Khan, M.T.; Rehaman, A.U.; Junaid, M.; Malik, S.I.; Wei, D.Q. Insight into novel clinical mutants of RpsA-S324F, E325K, and G341R of Mycobacterium tuberculosis associated with pyrazinamide resistance. Comput. Struct. Biotechnol. J. 2018, 16, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Junaid, M.; Khan, M.T.; Malik, S.I.; Wei, D.Q. Insights into the mechanisms of pyrazinamide resistance of three pyrazinamidase mutants N11K, P69T and D126N. J. Chem. Inf. Model. 2018. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Khan, M.T.; Liu, H.; Wadood, A.; Malik, S.I.; Chen, H.-F. Exploring the Pyrazinamide Drug Resistance Mechanism of Clinical Mutants T370P and W403G in Ribosomal Protein S1 of Mycobacterium tuberculosis. J. Chem. Inf. Model. 2019, 59, 1584–1597. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Malik, S.I.; Bhatti, A.I.; Ali, S.; Khan, A.S.; Zeb, M.T.; Nadeem, T.; Fazal, S. Pyrazinamide-resistant mycobacterium tuberculosis isolates from Khyber Pakhtunkhwa and rpsA mutations. J. Biol. Regul. Homeost. Agents 2018, 32, 705–709. [Google Scholar] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farah, S.I.; Abdelrahman, A.A.; North, E.J.; Chauhan, H. Opportunities and Challenges for Natural Products as Novel Antituberculosis Agents. Assay Drug Dev. Technol. 2016, 14, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future Med. Chem. 2011, 3, 1475–1489. [Google Scholar] [CrossRef]
- Hughes, C.C.; Fenical, W. Antibacterials from the Sea. Chemistry 2010, 16, 12512–12525. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.-M.; Wang, C.-Y.; Gerwick, W.H.; Shao, C.-L. Marine natural products as potential anti-tubercular agents. Eur. J. Med. Chem. 2019, 165, 273–292. [Google Scholar] [CrossRef]
- Felix, C.R.; Gupta, R.; Geden, S.; Roberts, J.; Winder, P.; Pomponi, S.A.; Diaz, M.C.; Reed, J.K.; Wright, A.E.; Rohde, K.H. Selective Killing of Dormant Mycobacterium tuberculosis by Marine Natural Products. Antimicrob. Agents Chemother. 2017, 61, e00743-17. [Google Scholar] [CrossRef]
- Zumla, A.; Nahid, P.; Cole, S.T. Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 2013, 12, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Dhar, N.; McKinney, J.; Manina, G. Phenotypic Heterogeneity in Mycobacterium tuberculosis. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Koul, A.; Arnoult, E.; Lounis, N.; Guillemont, J.; Andries, K. The challenge of new drug discovery for tuberculosis. Nature 2011, 469, 483–490. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, M.W.B.; Haltli, B.; Marchbank, D.H.; Kerr, R.G. Evaluation of pseudopteroxazole and pseudopterosin derivatives against Mycobacterium tuberculosis and other pathogens. Mar. Drugs 2012, 10, 1711–1728. [Google Scholar] [CrossRef] [PubMed]
- Canché Chay, C.I.; Gómez Cansino, R.; Espitia Pinzón, C.I.; Torres-Ochoa, R.O.; Martínez, R. Synthesis and anti-tuberculosis activity of the marine natural product caulerpin and its analogues. Mar. Drugs 2014, 12, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Martín, J.; Otero, L.; Palacios-Gutiérrez, J.J.; Fernández, J.; Mohamedi, Y.; Fontanil, T.; Salmón, M.; et al. Desertomycin G, a New Antibiotic with Activity against Mycobacterium tuberculosis and Human Breast Tumor Cell Lines Produced by Streptomyces althioticus MSM3, Isolated from the Cantabrian Sea Intertidal Macroalgae Ulva sp. Mar. Drugs 2019, 17, 114. [Google Scholar] [CrossRef] [PubMed]
- Bourguet-Kondracki, M.L.; Lacombe, F.; Guyot, M. Methanol adduct of puupehenone, a biologically active derivative from the marine sponge Hyrtios species. J. Nat. Prod. 1999, 62, 1304–1305. [Google Scholar] [CrossRef]
- Hagiwara, K.; Garcia Hernandez, J.E.; Harper, M.K.; Carroll, A.; Motti, C.A.; Awaya, J.; Nguyen, H.-Y.; Wright, A.D. Puupehenol, a potent antioxidant antimicrobial meroterpenoid from a Hawaiian deep-water Dactylospongia sp. sponge. J. Nat. Prod. 2015, 78, 325–329. [Google Scholar] [CrossRef]
- Gordaliza, M. Cytotoxic terpene quinones from marine sponges. Mar. Drugs 2010, 8, 2849–2870. [Google Scholar] [CrossRef]
- Ciavatta, M.L.; Lopez Gresa, M.P.; Gavagnin, M.; Romero, V.; Melck, D.; Manzo, E.; Guo, Y.-W.; van Soest, R.; Cimino, G. Studies on puupehenone-metabolites of a Dysidea sp.: Structure and biological activity. Tetrahedron 2007, 63, 1380–1384. [Google Scholar] [CrossRef]
- Robinson, S.J.; Hoobler, E.K.; Riener, M.; Loveridge, S.T.; Tenney, K.; Valeriote, F.A.; Holman, T.R.; Crews, P. Using enzyme assays to evaluate the structure and bioactivity of sponge-derived meroterpenes. J. Nat. Prod. 2009, 72, 1857–1863. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, E.A.; Yano, T.; Li, L.-S.; Avarbock, D.; Avarbock, A.; Helm, D.; McColm, A.A.; Duncan, K.; Lonsdale, J.T.; Rubin, H. Inhibitors of type II NADH: Menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. USA 2005, 102, 4548–4553. [Google Scholar] [CrossRef] [PubMed]
- Awasthy, D.; Ambady, A.; Narayana, A.; Morayya, S.; Sharma, U. Roles of the two type II NADH dehydrogenases in the survival of Mycobacterium tuberculosis in vitro. Gene 2014, 550, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.P.S.; Alonso, S.; Rand, L.; Dick, T.; Pethe, K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2008, 105, 11945–11950. [Google Scholar] [CrossRef] [PubMed]
- Kraus, G.A.; Nguyen, T.; Bae, J.; Hostetter, J.; Steadham, E. Synthesis and antitubercular activity of tricyclic analogs of puupehenone. Tetrahedron 2004, 60, 4223–4225. [Google Scholar] [CrossRef]
- Clark, R.J.; Field, K.L.; Charan, R.D.; Garson, M.J.; Brereton, M.; Willis, A.C. The haliclonacyclamines, cytotoxic tertiary alkaloids from the tropical marine sponge Haliclona sp. Tetrahedron 1998, 54, 8811–8826. [Google Scholar] [CrossRef]
- Arai, M.; Ishida, S.; Setiawan, A.; Kobayashi, M. Haliclonacyclamines, tetracyclic alkylpiperidine alkaloids, as anti-dormant mycobacterial substances from a marine sponge of Haliclona sp. Chem. Pharm. Bull. (Tokyo) 2009, 57, 1136–1138. [Google Scholar] [CrossRef]
- Jaspars, M.; Pasupathy, V.; Crews, P. A tetracyclic diamine alkaloid, halicyclamine A, from the marine sponge Haliclona sp. J. Org. Chem. 1994, 59, 3253–3255. [Google Scholar] [CrossRef]
- Arai, M.; Sobou, M.; Vilchéze, C.; Baughn, A.; Hashizume, H.; Pruksakorn, P.; Ishida, S.; Matsumoto, M.; Jacobs, W.R.; Kobayashi, M. Halicyclamine A, a marine spongean alkaloid as a lead for anti-tuberculosis agent. Bioorg. Med. Chem. 2008, 16, 6732–6736. [Google Scholar] [CrossRef]
- Garson, M.J.; Flowers, A.E.; Webb, R.I.; Charan, R.D.; McCaffrey, E.J. A sponge/dinoflagellate association in the haplosclerid sponge Haliclona sp.: Cellular origin of cytotoxic alkaloids by percoll density gradient fractionation. Cell Tissue Res. 1998, 293, 365–373. [Google Scholar] [CrossRef]
- Wei, X.; Nieves, K.; Rodríguez, A.D. Neopetrosiamine A, biologically active bis-piperidine alkaloid from the Caribbean sea sponge Neopetrosia proxima. Bioorg. Med. Chem. Lett. 2010, 20, 5905–5908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manganelli, R.; Voskuil, M.I.; Schoolnik, G.K.; Dubnau, E.; Gomez, M.; Smith, I. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 2002, 45, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef]
- Ghosh, T.; Bose, D.; Zhang, X. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 2010, 34, 611–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuorre, M. Quantitative Literature Review with R: Exploring Psychonomic Society Journals, Part I. Available online: https://vuorre.netlify.com/post/2017/quantitative-literature-review-with-r-part-i/ (accessed on 9 October 2018).
- Rani, J.; Shah, A.B.R.; Ramachandran, S. pubmed.mineR: An R package with text-mining algorithms to analyse PubMed abstracts. J. Biosci. 2015, 40, 671–682. [Google Scholar] [CrossRef]
- RISmed: Download Content from NCBI Databases Version 2.1.7 from CRAN. Available online: https://rdrr.io/cran/RISmed/ (accessed on 9 October 2018).
- Larsen, K.G.; Mikucionis, M.; Nielsen, B.; Skou, A. Testing Real-time Embedded Software Using UPPAAL-TRON: An Industrial Case Study. In Proceedings of the 5th ACM International Conference on Embedded Software 2005, Jersey City, NJ, USA, 18–22 September 2005; pp. 299–306. [Google Scholar]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-Controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ritz, A.; Poirel, C.L.; Tegge, A.N.; Sharp, N.; Simmons, K.; Powell, A.; Kale, S.D.; Murali, T. Pathways on demand: Automated reconstruction of human signaling networks. NPJ Syst. Biol. Appl. 2016, 2, 16002. [Google Scholar] [CrossRef]
- Rodríguez, Y.; Alejo, C.; Alejo, I.; Viguria, A. ANIMO, framework to simplify the real-time distributed communication. In Management of Cyber Physical Objects in the Future Internet of Things; Guerrieri, A., Loscri, V., Rovella, A., Fortino, G., Eds.; Springer: Berlin, Germany, 2016; pp. 77–92. [Google Scholar]
- Kaushik, A.C. Logisim Operon Circuits. IJSER 2013, 4, 5. [Google Scholar]




| Compound | Formula | Molecular Mass (kDa) | MICR (g/mL) a | MICD (g/mL) b | MICR/MICD | IC50 (g/mL) | SIR c | SID d | Source |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C31H30Br6N4O11 | 1,114.02 | 8.5 | Inactive | NA | 200 | 23.5 | NA | HBOI.047.F07 |
| 2 | C19H40O3 | 316.53 | 60.8 | 22.5 | 2.7 | 200 | 3.3 | 8.5 | HBOI.047.F07 |
| 3 | C16H30O2 | 254.41 | 28.5 | 7.9 | 3.6 | 200 | 7.0 | 31.1 | HBOI.031.C02 |
| 4 | C22H32O4 | 360.49 | 11.3 | 0.5 | 21.8 | 8 | 0.7 | 15.5 | HBOI.050.F04 |
| 5 | C21H26O3 | 326.44 | 87.6 | 15.4 | 5.6 | 50.4 | 0.6 | 6.2 | HBOI.050.F04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.T.; Kaushik, A.C.; Bhatti, A.I.; Zhang, Y.-J.; Zhang, S.; Wei, A.J.; Malik, S.I.; Wei, D.Q. Marine Natural Products and Drug Resistance in Latent Tuberculosis. Mar. Drugs 2019, 17, 549. https://doi.org/10.3390/md17100549
Khan MT, Kaushik AC, Bhatti AI, Zhang Y-J, Zhang S, Wei AJ, Malik SI, Wei DQ. Marine Natural Products and Drug Resistance in Latent Tuberculosis. Marine Drugs. 2019; 17(10):549. https://doi.org/10.3390/md17100549
Chicago/Turabian StyleKhan, Muhammad Tahir, Aman Chandra Kaushik, Aamer Iqbal Bhatti, Yu-Juan Zhang, Shulin Zhang, Amie Jinghua Wei, Shaukat Iqbal Malik, and Dong Qing Wei. 2019. "Marine Natural Products and Drug Resistance in Latent Tuberculosis" Marine Drugs 17, no. 10: 549. https://doi.org/10.3390/md17100549
APA StyleKhan, M. T., Kaushik, A. C., Bhatti, A. I., Zhang, Y. -J., Zhang, S., Wei, A. J., Malik, S. I., & Wei, D. Q. (2019). Marine Natural Products and Drug Resistance in Latent Tuberculosis. Marine Drugs, 17(10), 549. https://doi.org/10.3390/md17100549