Short Chain Fatty Acid Biosynthesis in Microalgae Synechococcus sp. PCC 7942
Abstract
:1. Introduction
2. Results and Discussion
2.1. Fatty Acid Compositions of Synechococcus sp. PCC7942 under Different Conditions
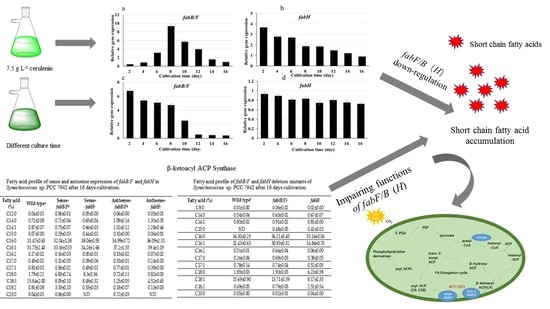
2.2. Genes Expression under Different Conditions in Synechococcus sp. PCC7942
2.3. Fatty Acid Compositions in fabB/F and fabH Sense/Antisense Expression Strains
2.4. Fatty Acid Compositions of fabB/F and fabH Deletion Mutants
2.5. Complementation of E. coli
2.6. Fluorescence Localization of the Related Proteins
3. Materials and Methods
3.1. Microalgae Cultures
3.2. Genomic DNA Extraction and PCR Analysis
3.3. Sense and Antisense Expression Vector Construction
3.4. Deletion Mutant Construction
3.5. Fusion Expression Vector of Green Fluorescence Protein Construction
3.6. Liquid Culture and Screening of Transgenic Microalgae
3.7. RNA Extraction and cDNA Synthesis
3.8. Quantitative Real-Time PCR
3.9. Lipid Extraction and Fatty Acid Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Traul, K.A.; Driedger, A.; Ingle, D.L.; Nakhasi, D. Review of the toxicologic properties of medium-chain triglycerides. Food Chem. Toxicol. 2000, 38, 79–98. [Google Scholar] [CrossRef]
- Cournarie, F.; Savelli, M.P.; Rosilio, V.; Bretez, F.; Vauthier, C.; Grossiord, J.L.; Seiller, M. Insulin-loaded W/O/W multiple emulsions: Comparison of the performances of systems prepared with medium-chain-triglycerides and fish oil. Eur. J. Pharm. Biopharm. 2004, 58, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Dehesh, K. How can we genetically engineer oilseed crops to produce high levels of medium-chain fatty acids? Eur. J. Lipid Sci. Technol. 2015, 103, 688–697. [Google Scholar] [CrossRef]
- Xia, H.; Wang, X.; Li, M.; Xiao, H. Improving fatty acid composition and increasing triacylglycerol content in plants by gene engineering. Chin. J. Biotech. 2010, 26, 735–743. [Google Scholar]
- Shi, T.T. Screening of Oil-producing Strains; Shandong Polytechnic University: Jinan, China, 2012. [Google Scholar]
- Schenk, P.M.; Thomas-Hall, S.R.; Stephens, E.; Marx, U.C.; Mussgnug, J.H.; Posten, C.; Kruse, O.; Hankamer, B. Second Generation Biofiiels; High-Efficiency Microalgae for Biodiesel Production. Bioenerg. Res. 2008, 1, 20–43. [Google Scholar] [CrossRef]
- Xu, L.; Li, Z.; Zhou, H.L.; Ding, Y.T.; Liu, L. Study of hygroscopic and moisturizing performance of oligosaccharides obtained from enzymolysis of algin. China Surfactant Deterg. Cosmet. 2011, 41, 42–45. [Google Scholar]
- Lü, H.; Gao, Y.; Shan, H.; Lin, Y. Preparation and antibacterial activity studies of degraded polysaccharide selenide from Enteromorpha prolifera. Carbohydr. Polym. 2014, 107, 98–102. [Google Scholar] [CrossRef]
- Abad, M.J.; Bedoya, L.M.; Bermejo, P. Natural marine anti-inflammatory products. Mini Rev. Med. Chem. 2008, 8, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chung, D.; Shin, I.S.; Lee, H.; Kim, J.; Lee, Y.; You, S. Effects of molecular weight and hydrolysis conditions on anticancer activity of fucoidans from sporophyll of Undaria pinnatifida. Int. J. Biol. Macromol. 2008, 43, 433–437. [Google Scholar] [CrossRef]
- Shao, P.; Chen, X.; Sun, P. Chemical characterization, antioxidant and antitumor activity of sulfated polysaccharide from Sargassum horneri. Carbohydr. Polym. 2014, 105, 260–269. [Google Scholar] [CrossRef]
- Adams, N.L.; Shick, J.M. Mycosporine-like amino acids provide protection against ultraviolet radiation in eggs of the green sea urchin Strongylocentrotus droebachiensis. Photochem. Photobiol. 1996, 64, 149–158. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, N.; Zhou, G.; Lu, X.; Xu, Z.; Li, Z. In vivo antioxidant activity of polysaccharide fraction from Porphyra haitanesis (Rhodephyta) in aging mice. Pharmacol. Res. 2003, 48, 151–155. [Google Scholar] [CrossRef]
- Abidov, M.; Ramazanov, Z.; Seifulla, R.; Grachev, S. The effects of Xanthigen in the weight management of obese premenopausal women with non-alcoholic fatty liver disease and normal liver fat. Diabetes Obes. Metab. 2010, 12, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.C. Application of Bioengineering in Olechemical Industry (III)—Application of Medium Chain Length Fatty Acid Esters in Medical. Nutr. Cosmet. China Oils Fats 1999, 6, 50–52. [Google Scholar] [CrossRef]
- Kim, I.H.; Kim, H.; Lee, K.T.; Chung, S.H.; Ko, S.N. Lipase-catalyzed acidolysis of perilla oil with caprylic acid to produce structured lipids. J. Am. Oil Chem. Soc. 2002, 79, 363–367. [Google Scholar] [CrossRef]
- Murga, M.L.; Cabrera, G.M.; De Valdez, G.F.; Disalvo, A.; Seldes, A.M. Influence of growth temperature on cryotolerance and lipid composition of Lactobacillus acidophilus. J. Appl. Microbiol. 2000, 88, 342–348. [Google Scholar] [CrossRef]
- Yumoto, I.; Yamazaki, K.; Hishinuma, M.; Nodasaka, Y.; Inoue, N.; Kawasaki, K. Identification of facultatively alkaliphilic Bacillus sp. strain YN-2000 and its fatty acid composition and cell-surface aspects depending on culture pH. Extremophiles 2000, 4, 285–290. [Google Scholar] [CrossRef]
- Hartig, C.; Loffhagen, N.; Babel, W. Glucose stimulates a decrease of the fatty acid saturation degree in Acinetobacter calcoaceticus. Arch. Microbiol. 1999, 171, 166–172. [Google Scholar] [CrossRef]
- Huang, J.Z.; Aki, T.; Hachida, K.; Yokochi, T.; Kawamoto, S.; Shigeta, S.; Ono, K.; Suzuki, O. Profile of Polyunsaturated Fatty Acids Produced by Thraustochytrium sp. KK17-3. J. Am. Oil Chem. Soc. 2001, 78, 605–610. [Google Scholar] [CrossRef]
- Joestensen, J.P.; Landfald, B. Influence of growth conditions on fatty acid composition of a polyunsaturated-fatty-acid-producing Vibrio species. Arch. Microbiol. 1996, 165, 306–310. [Google Scholar] [CrossRef]
- Han, J.C. Research on the Collection and Lipid Systhesis Pathway of Oil-Producing Microalgae; Ocean University of China: Qingdao, China, 2012. [Google Scholar]
- Jones, A.; Davies, H.M.; Voelker, T.A. Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell 1995, 7, 359–371. [Google Scholar] [CrossRef]
- Dehesh, K.; Jones, A.; Knutzon, D.S.; Voelker, T.A. Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of ChFatB2, a thioesterase cDNA from Cuphea hookeriana. Plant J. 1996, 2, 167–172. [Google Scholar] [CrossRef]
- Thelen, J.J.; Ohlrogge, J.B. Metabolic engineering of fatty acid biosynthesis in plants. Metab. Eng. 2002, 4, 12–21. [Google Scholar] [CrossRef]
- Lu, S.F. Biosynthesis and Gene Engineering of Plant Fatty Acids. Chin. Bull. Bot. 2000, 17, 481–491. [Google Scholar]
- Li, X.W. Cloning and Analysis of Genes of Fatty Acid Biosynthesis-related Enzymes (KASI, FatB) and Genetic Transformation of ahFAD2B in Arachis hypogaea L.; Shandong Agricultural University: Taian, China, 2011. [Google Scholar]
- Verwoert, I.I.; van der Linder, K.H.; Walsh, M.C. Modification of Brassica napusseed oil by expression of the Escherichia coli fabH gene, encoding β-ketoacyl-acyl carrier protein synthase III. Plant Mol. Biol. 1995, 27, 875–886. [Google Scholar] [CrossRef]
- Dehesh, K.; Tai, H.; Edwards, P.; Byrne, J.; Jaworski, J.G. Overexpression of β-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001, 125, 1103–1114. [Google Scholar] [CrossRef]
- Kuo, J.; Khosla, C. The initiation keto synthase (FabH) is the sole rate-limiting enzyme of the fatty acid synthase of Synechococcus sp. PCC 7002. Metab. Eng. 2014, 22, 53–59. [Google Scholar] [CrossRef]
- Renaud, S.M.; Thinh, L.V.; Lambrinidis, G.; Parry, D.L. Effect of temperature on growth, chemical composition and fatty acid composition of tropical Australian microalgae grown in batch cultures. Aquaculture 2002, 211, 195–214. [Google Scholar] [CrossRef]
- Parker, P.L.; van Baalen, C.; Maurer, L. Fatty acids in eleven species of blue-green algae: Geochemical significance. Science 1967, 155, 707–708. [Google Scholar] [CrossRef] [PubMed]
- Karatay, S.E.; Dönmez, G. Microbial oil production from thermophile cyanobacteria for biodiesel production. Appl. Energ. 2011, 88, 3632–3635. [Google Scholar] [CrossRef]
- Arnvig, M.K.; McGuire, J.N.; von Wettstein-Knowles, P. Acyl carrier protein (ACP) inhibition and other differences between β-ketoacyl synthase (KAS) I and II. Biochem. Soc. Trans. 2000, 28, 607–610. [Google Scholar] [CrossRef]
- Jin, L.M.; Liu, P.; Tian, W.J. Bacterial fatty acid biosynthesis enzymes-drug targets for antibacterial agent screen. J. Shenyang Pharm. Univ. 2006, 23, 814–818. [Google Scholar]
- Israel, G.; James, R.; Konrad, B. Inhibition of Lipid Synthesis in Escherichia coli Cells by the Antibiotic cerulenin. Antimicrob. Agents Chemother. 1973, 5, 549–554. [Google Scholar]
- Lin, P.H. Preliminary Modeling Screening for Fatty Acid Synthase FabB/F Inhibitor; Jiangxi Agricultural University: Nanchang, China, 2012. [Google Scholar]
- Blatti, J.L.; Beld, J.; Behnke, C.A.; Mendez, M.; Mayfield, S.P.; Burkart, M.D. Manipulating fatty acid biosynthesis in microalgae for biofuel through protein-protein interactions. PLoS ONE 2012, 7, e42949. [Google Scholar] [CrossRef]
- Ter Beek, A.; Keijser, B.J.; Boorsma, A.; Zakrzewska, A.; Orij, R.; Smits, G.J.; Brul, S. Transcriptome analysis of sorbic acid-stressed Bacillus subtilis reveals a nutrient limitation response and indicates plasma membrane remodeling. J. Bacteriol. 2008, 190, 1751–1761. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.H.; Ma, J.R.; Miao, X.Y.; Wang, H.H. Biological Function Research of β-ketoacyl ACP Synthase III. Prog. Biochem. Biophys. 2016, 43, 1004–1012. [Google Scholar]
- Chen, J.Q.; Lang, C.X.; Hu, Z.H. Antisense PEP Gene Regulates to Ratio of Protein and Lipid Content in Brassica Napus Seeds. J. Agric. Biotechnol. 1999, 7, 316–320. [Google Scholar]
- Song, D.; Hou, L.; Shi, D. Exploitation and utilization of rich lipids-microalgae, as new lipids feedstock for biodiesel production—A review. Chin. J. Biotechnol. 2008, 24, 341–348. [Google Scholar] [CrossRef]
- Lin, H.X.; Lee, Y.K. Genetic engineering of medium-chain-length fatty acid synthesis in Dunaliella tertiolecta for improved biodiesel production. J. Appl. Phycol. 2017, 29, 2811–2819. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.C.; Deng, L.T.; Tong, W.H.; Zhu, L.; Wang, H.H. Identification and Function Reasearch of Five β-Ketoacyl-ACP Synthase Homologues. Prog. Biochem. Biophys. 1997, 41, 887–895. [Google Scholar]
- Qian, W.Z. Preparation of Monoclonal Antibody against Protein FabB and Analysis of the Difference and Distribution of FabB in Different Bactieria Species; Yangzhou University: Yangzhou, China, 2013. [Google Scholar]
- Leonard, J.; Knapp, S.J.; Slabaugh, M.B. A Cuphea β-ketoacyl ACP synthase shifts the synthesis of fatty acids towards shorter chains in Arabidopsis seeds expressing Cuphea FatB thioesterases. Plant J. 1998, 13, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Pidkowich, M.S.; Nguyen, H.T.; Heilmann, I.; Ischebeck, T.; Shanklin, J. Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc. Natl. Acad. Sci. USA 2007, 104, 4742–4747. [Google Scholar] [CrossRef]
- González-Mellado, D.; von Wettstein-Knowles, P.; Garcés, R.; Martínez-Force, E. The role of β-ketoacyl-acyl carrier protein synthase III in the condensation steps of fatty acid biosynthesis in sunflower. Planta 2010, 231, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Brück, F.M.; Brummel, M.; Schuch, R.; Spener, F. In-vitro evidence for feed-back regulation of beta-ketoacyl-acyl carrier protein synthase III in medium-chain fatty acid biosynthesis. Planta 1996, 198, 271–278. [Google Scholar] [CrossRef]
- Post-Beittenmiller, D.; JaworskiJ, G.; Ohlrogge, J.B. In vivo pools of free and acylated acyl carrier proteins in spinach. Evidence for Sites of regulation of fatty acid biosynthesis. J. Biol. Chem. 1991, 266, 1858–1865. [Google Scholar] [PubMed]
- Roessler, P.G. Changes in the activities of various lipid and carbohydrate biosynthetic enzymes in the diatom Cyclotella cryptica in response to silicon deficiency. Arch. Biochem. Biophys. 1988, 267, 521–528. [Google Scholar] [CrossRef]
- Courchesne, N.M.; Parisien, A.; Wang, B.; Lan, C.Q. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 2009, 141, 31–41. [Google Scholar] [CrossRef]
- Perrineau, M.M.; Zelzion, E.; Gross, J.; Price, D.C.; Boyd, J.; Bhattacharya, D. Evolution of salt tolerance in a laboratory reared population of Chlamydomonas reinhardtii. Environ. Microbiol. 2014, 16, 1755–1766. [Google Scholar] [CrossRef] [PubMed]



| Fatty Acid (%) | Time (day) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | 20 | |
| 12:0 | 0.02 ± 0.01 | ND | 0.02 ± 0.00 | ND | ND | ND | ND | ND | ND | ND |
| 14:0 | 0.63 ± 0.03 | 0.74 ± 0.15 | 0.56 ± 0.06 | 0.45 ± 0.03 | 0.62 ± 0.03 | 0.27 ± 0.03 | 0.33 ± 0.01 | 0.42 ± 0.03 | 0.34 ± 0.03 | 0.36 ± 0.03 |
| 14:1 | 0.98 ± 0.02 | 1.70 ± 0.19 | 1.17 ± 0.19 | 0.87 ± 0.10 | 0.90 ± 0.06 | 0.48 ± 0.03 | 0.55 ± 0.04 | 0.67 ± 0.06 | 0.57 ± 0.03 | 0.60 ± 0.06 |
| 16:0 | 42.14 ± 3.96 | 39.98 ± 2.67 | 45.42 ± 2.64 | 41.71 ± 2.22 | 38.65 ± 2.68 | 47.93 ± 2.00 | 45.41 ± 2.11 | 46.56 ± 2.04 | 46.63 ± 3.08 | 46.20 ± 2.05 |
| 16:1 | 34.72 ± 2.04 | 38.20 ± 1.89 | 36.80 ± 1.74 | 31.03 ± 1.25 | 31.32 ± 0.71 | 26.96 ± 1.71 | 31.94 ± 1.87 | 29.89 ± 0.89 | 29.60 ± 1.60 | 30.48 ± 1.89 |
| 16:2 | ND | ND | ND | ND | ND | ND | ND | ND | 0.13 ± 0.03 | 0.13 ± 0.02 |
| 17:0 | 0.13 ± 0.03 | 0.22 ± 0.03 | 0.09 ± 0.03 | 0.12 ± 0.03 | 0.11 ± 0.02 | 0.18 ± 0.04 | 0.11 ± 0.03 | 0.10 ± 0.00 | 0.13 ± 0.01 | 0.11 ± 0.00 |
| 17:1 | 0.22 ± 0.01 | 0.41 ± 0.01 | 0.14 ± 0.03 | 0.24 ± 0.05 | 0.27 ± 0.02 | 0.30 ± 0.00 | 0.33 ± 0.00 | 0.34 ± 0.03 | 0.32 ± 0.01 | 0.33 ± 0.03 |
| 18:0 | 1.70 ± 0.34 | 3.00 ± 0.21 | 0.24 ± 0.05 | 2.49 ± 0.24 | 3.11 ± 0.25 | 2.57 ± 0.25 | 1.89 ± 0.37 | 1.08 ± 0.11 | 1.85 ± 0.11 | 1.61 ± 0.03 |
| 18:1 | 2.51 ± 0.33 | 3.89 ± 0.29 | 1.80 ± 0.08 | 3.79 ± 0.11 | 4.48 ± 0.31 | 7.14 ± 0.75 | 6.42 ± 0.57 | 4.90 ± 0.36 | 6.15 ± 0.36 | 5.82 ± 0.23 |
| 18:2 | 0.31 ± 0.047 | 0.24 ± 0.09 | 3.16 ± 0.32 | 0.34 ± 0.04 | 0.39 ± 0.03 | 0.35 ± 0.02 | 0.68 ± 0.03 | 0.21 ± 0.03 | 0.41 ± 0.02 | 0.43 ± 0.03 |
| 20:0 | 0.04 ± 0.00 | 0.03 ± 0.00 | 0.04 ± 0.00 | 0.08 ± 0.03 | 0.12 ± 0.01 | 0.05 ± 0.01 | ND | ND | 0.06 ± 0.01 | 0.13 ± 0.02 |
| 22:0 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.05 ± 0.01 | ND | 0.19 ± 0.01 | ND | ND | ND | 0.26 ± 0.02 | 0.27 ± 0.04 |
| 22:1 | 0.05 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 | ND | 0.11 ± 0.01 | 0.10 ± 0.00 | 0.07 ± 0.00 | ND | ND | 0.06 ± 0.00 |
| Fatty Acid (%) | Cerulenin Concentration (g/L) | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2.5 | 5 | 7.5 | |
| 12:0 | 0.04 ± 0.00 | 0.05 ± 0.00 | 0.04 ± 0.01 | 0.07 ± 0.00 | 0.09 ± 0.01 |
| 14:0 | 0.71 ± 0.08 | 0.98 ± 0.09 | 0.77 ± 0.04 | 0.84 ± 0.09 | 1.25 ± 0.31 |
| 14:1 | 0.87 ± 0.15 | 1.1 ± 0.26 | 0.82 ± 0.08 | 1.35 ± 0.17 | 1.5 ± 0.31 |
| 15:0 | 0.07 ± 0.02 | 0.07 ± 0.01 | ND | ND | ND |
| 16:0 | 31.47 ± 2.48 | 41.74 ± 1.85 | 34.76 ± 1.68 | 35.43 ± 1.53 | 31.84 ± 2.46 |
| 16:1 | 31.75 ± 2.39 | 32.97 ± 2.56 | 25.66 ± 2.90 | 26.57 ± 3.38 | 25.43 ± 2.45 |
| 16:2 | 0.27 ± 0.03 | 0.07 ± 0.02 | ND | 0.06 ± 0.01 | 0.08 ± 0.01 |
| 17:0 | 0.49 ± 0.03 | 0.16 ± 0.04 | 0.16 ± 0.02 | 0.21 ± 0.02 | 0.47 ± 0.01 |
| 17:1 | 0.81 ± 0.12 | 0.43 ± 0.02 | 0.52 ± 0.06 | 0.51 ± 0.04 | 0.76 ± 0.11 |
| 18:0 | 1.79 ± 0.10 | 2.14 ± 0.42 | 3.33 ± 0.33 | 2.7 ± 0.15 | 3.57 ± 0.26 |
| 18:1 | 15.84 ± 0.98 | 8.02 ± 0.14 | 14.05 ± 1.47 | 12.13 ± 1.49 | 5.69 ± 0.04 |
| 18:2 | 0.81 ± 0.08 | 1.15 ± 0.11 | 0.69 ± 0.03 | 0.9 ± 0.10 | 4.44 ± 0.27 |
| 20:0 | 0.04 ± 0.03 | 0.05 ± 0.00 | ND | ND | ND |
| Fatty Acid (%) | Wild Type a | Sense-fabB/F b | Sense-fabH c | Antisense-fabB/F d | Antisense-fabH e |
|---|---|---|---|---|---|
| C12:0 | 0.04 ± 0.01 | 0.06 ± 0.01 | 0.05 ± 0.00 | 0.06 ± 0.00 | 0.05 ± 0.01 |
| C14:0 | 0.71 ± 0.08 | 0.71 ± 0.06 | 0.65 ± 0.04 | 1.09 ± 0.16 | 1.35 ± 0.35 |
| C14:1 | 0.87 ± 0.07 | 0.75 ± 0.07 | 0.66 ± 0.03 | 1.52 ± 0.12 | 2.28 ± 0.46 |
| C15:0 | 0.07 ± 0.00 | 0.29 ± 0.03 | 0.44 ± 0.02 | 0.10 ± 0.00 | 0.06 ± 0.01 |
| C16:0 | 31.47 ± 0.63 | 32.04 ± 1.38 | 38.06 ± 0.55 | 34.99 ± 0.72 | 36.09 ± 2.31 |
| C16:1 | 31.75 ± 2.40 | 33.16 ± 0.53 | 34.26 ± 1.68 | 37.2 ± 1.35 | 39.4 ± 1.19 |
| C16:2 | 0.27 ± 0.02 | 0.16 ± 0.03 | 0.05 ± 0.01 | 0.13 ± 0.02 | 0.07 ± 0.02 |
| C17:0 | 0.49 ± 0.02 | 0.32 ± 0.05 | 0.39 ± 0.06 | 0.10 ± 0.01 | 0.24 ± 0.03 |
| C17:1 | 0.81 ± 0.01 | 0.56 ± 0.02 | 0.49 ± 0.02 | 0.77 ± 0.01 | 0.39 ± 0.03 |
| C18:0 | 1.79 ± 0.21 | 6.83 ± 0.74 | 8.3 ± 0.36 | 0.72 ± 0.11 | 0.82 ± 0.03 |
| C18:1 | 15.84 ± 2.00 | 8.05 ± 0.10 | 8.49 ± 0.32 | 1.25 ± 0.05 | 4.52 ± 0.65 |
| C18:2 | 0.81 ± 0.08 | 3.33 ± 0.20 | 0.33 ± 0.03 | 0.18 ± 0.07 | 0.13 ± 0.03 |
| C20:0 | 0.04 ± 0.01 | 0.06 ± 0.00 | ND | 0.72 ± 0.03 | ND |
| Fatty Acid (%) | Wild Type a | fab(B/F)− | fabH− |
|---|---|---|---|
| C8:0 | 0.01 ± 0.00 | 0.06 ± 0.02 | 0.02 ± 0.00 |
| C14:0 | 0.54 ± 0.04 | 0.63 ± 0.02 | 0.67 ± 0.07 |
| C14:1 | 0.85 ± 0.02 | 0.91 ± 0.02 | 0.81 ± 0.00 |
| C15:0 | ND | 0.48 ± 0.05 | 0.41 ± 0.03 |
| C16:0 | 34.30 ± 0.25 | 36.21 ± 0.40 | 33.16 ± 0.04 |
| C16:1 | 32.43 ± 0.63 | 30.93 ± 0.32 | 34.86 ± 0.70 |
| C16:2 | 0.51 ± 0.01 | 0.64 ± 0.04 | 0.06 ± 0.00 |
| C17:0 | 0.26 ± 0.06 | 0.69 ± 0.03 | 0.38 ± 0.05 |
| C17:1 | 0.78 ± 0.14 | 0.74 ± 0.04 | 0.52 ± 0.05 |
| C18:0 | 1.85 ± 0.03 | 1.93 ± 0.03 | 6.23 ± 0.58 |
| C18:1 | 15.69 ± 0.90 | 13.71 ± 0.39 | 8.17 ± 0.35 |
| C18:2 | 0.69 ± 0.05 | 0.79 ± 0.03 | 2.51 ± 0.54 |
| C20:0 | 0.05 ± 0.00 | 0.02 ± 0.01 | 0.04 ± 0.00 |
| Fatty Acid (%) | E. coli BL21 | fabB− | fabF− | fabH− | Com-fabB a | Com-fabF b | Com-fabH c |
|---|---|---|---|---|---|---|---|
| C12:0 | 0.23 ± 0.02 | 0.3 ± 0.00 | 0.72 ± 0.10 | 0.78 ± 0.13 | 0.31 ± 0.12 | 0.50 ± 0.10 | 0.23 ± 0.01 |
| C13:0 | 1.33 ± 0.11 | ND | ND | 0.10 ± 0.00 | 1.19 ± 0.00 | 0.94 ± 0.23 | 1.87 ± 0.34 |
| C14:0 | 2.88 ± 0.28 | 3.04 ± 0.23 | 3.69 ± 0.29 | 3.01 ± 0.46 | 2.88 ± 0.46 | 3.03 ± 0.33 | 3.87 ± 0.58 |
| C14:1 | ND | 3.23 ± 0.20 | 0.21 ± 0.01 | 0.30 ± 0.00 | ND | ND | ND |
| C15:0 | 8.25 ± 0.22 | 0.57 ± 0.03 | 1.02 ± 0.07 | 0.72 ± 0.15 | 6.32 ± 0.15 | 5.97 ± 0.55 | 12.45 ± 2.02 |
| C16:0 | 33.98 ± 3.83 | 50.51 ± 2.75 | 48.59 ± 3.11 | 50.23 ± 3.64 | 33.82 ± 0.64 | 33.37 ± 2.50 | 34.76 ± 1.55 |
| C16:1 | 2.78 ± 0.30 | 9.21 ± 0.22 | 9.94 ± 1.60 | 8.21 ± 0.94 | 2.60 ± 0.94 | 2.66 ± 0.35 | 3.08 ± 0.19 |
| C17:0 | 5.02 ± 0.21 | 0.44 ± 0.04 | 0.60 ± 0.18 | 0.51 ± 0.07 | 3.56 ± 0.07 | 3.94 ± 0.42 | 7.57 ± 0.67 |
| C17:1 | 2.69 ± 0.17 | 2.97 ± 0.31 | 2.46 ± 0.38 | 5.18 ± 0.56 | 3.71 ± 0.56 | 2.36 ± 0.34 | 1.99 ± 0.12 |
| C18:0 | 2.30 ± 0.16 | 1.40 ± 0.15 | 2.93 ± 0.54 | 2.37 ± 0.40 | 2.62 ± 0.40 | 2.07 ± 0.23 | 2.20 ± 0.24 |
| C18:1 | 20.57 ± 0.94 | 6.61 ± 0.51 | 7.12 ± 0.29 | 5.39 ± 0.68 | 20.51 ± 0.68 | 20.62 ± 2.83 | 5.55 ± 0.77 |
| C18:2 | 0.17 ± 0.02 | 0.34 ± 0.02 | 0.52 ± 0.02 | 0.37 ± 0.03 | 0.16 ± 0.03 | 0.17 ± 0.03 | 8.36 ± 0.51 |
| C19:0 | 1.11 ± 0.11 | 1.34 ± 0.05 | 1.22 ± 0.12 | 2.83 ± 0.29 | 5.20 ± 0.29 | 1.25 ± 0.15 | 0.96 ± 0.14 |
| C20:0 | 0.03 ± 0.01 | 0.02 ± 0.00 | 0.04 ± 0.01 | 0.03 ± 0.00 | 0.04 ± 0.00 | 0.03 ± 0.00 | 0.03 ± 0.01 |
| C20:1 | 1.59 ± 0.17 | 0.41 ± 0.04 | 0.73 ± 0.03 | 0.31 ± 0.03 | 1.76 ± 0.03 | 1.98 ± 0.17 | 1.03 ± 0.03 |
| C21:0 | 0.01 ± 0.00 | ND | 0.02 ± 0.00 | 0.02 ± 0.01 | 0.01 ± 0.01 | ND | ND |
| C22:0 | 0.12 ± 0.01 | 0.03 ± 0.00 | 0.14 ± 0.02 | 0.07 ± 0.01 | 0.11 ± 0.01 | 0.09 ± 0.02 | 0.16 ± 0.01 |
| C22:1 | 0.05 ± 0.00 | 0.02 ± 0.01 | ND | ND | 0.08 ± 0.03 | 0.02 ± 0.00 | ND |
| C23:0 | 0.10 ± 0.00 | 0.03 ± 0.01 | 0.05 ± 0.00 | 0.04 ± 0.01 | 0.06 ± 0.01 | 0.07 ± 0.00 | 0.17 ± 0.00 |
| C24:0 | 0.09 ± 0.00 | 0.02 ± 0.00 | 0.05 ± 0.00 | 0.04 ± 0.01 | 0.06 ± 0.01 | 0.05 ± 0.00 | 0.15 ± 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, Y.; Miao, X. Short Chain Fatty Acid Biosynthesis in Microalgae Synechococcus sp. PCC 7942. Mar. Drugs 2019, 17, 255. https://doi.org/10.3390/md17050255
Gong Y, Miao X. Short Chain Fatty Acid Biosynthesis in Microalgae Synechococcus sp. PCC 7942. Marine Drugs. 2019; 17(5):255. https://doi.org/10.3390/md17050255
Chicago/Turabian StyleGong, Yi, and Xiaoling Miao. 2019. "Short Chain Fatty Acid Biosynthesis in Microalgae Synechococcus sp. PCC 7942" Marine Drugs 17, no. 5: 255. https://doi.org/10.3390/md17050255
APA StyleGong, Y., & Miao, X. (2019). Short Chain Fatty Acid Biosynthesis in Microalgae Synechococcus sp. PCC 7942. Marine Drugs, 17(5), 255. https://doi.org/10.3390/md17050255