Structure Elucidation and Functional Studies of a Novel β-hairpin Antimicrobial Peptide from the Marine Polychaeta Capitella teleta
Abstract
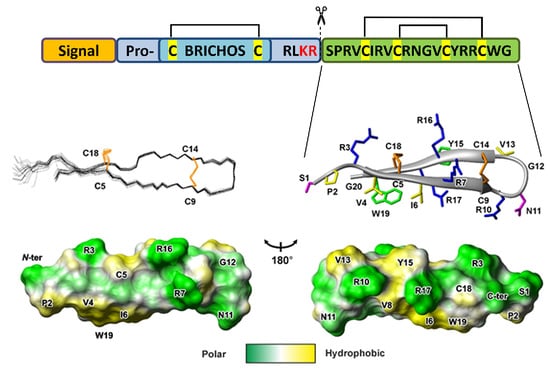
:1. Introduction
2. Results and Discussion
2.1. Recombinant Production of the Capitellacin
2.2. Spatial Structure of the Capitellacin in Aqueous Solution
2.3. Capitellacin Interaction with Model Membranes
2.4. Conformation of Capitellacin in Membrane-mimicking Detergent Micelles
2.5. Proton Transfer Activity of Capitellacin
2.6. Biological Activity and Mechanism of Antibacterial Action
3. Materials and Methods
3.1. Recombinant Production of the Peptides
3.2. NMR Spectroscopy and 3D Structure Calculation
3.3. Accessing Codes
3.4. Preparation of the Peptide-Containing Small Unilamellar Vesicles
3.5. Circular Dichroism Spectroscopy
3.6. Fourier-Transform Infrared Spectroscopy
3.7. Tryptophan Fluorescence and Quenching
3.8. Proton Transport Measurements
3.9. Antimicrobial Assays
3.10. Bacterial Membranes Permeability Assay
3.11. Hemolysis and Cytotoxicity Assay
3.12. Cell-Free Protein Expression Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V.; Aleshina, G.M.; Balandin, S.V.; Krasnosdembskaya, A.D.; Markelov, M.L.; Frolova, E.I.; Leonova, Y.F.; Tagaev, A.A.; Krasnodembsky, E.G.; Kokryakov, V.N. Purification and primary structure of two isoforms of arenicin, a novel antimicrobial peptide from marine polychaeta Arenicola marina. FEBS Lett. 2004, 577, 209–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tasiemski, A.; Jung, S.; Boidin-Wichlacz, C.; Jollivet, D.; Cuvillier-Hot, V.; Pradillon, F.; Vetriani, C.; Hecht, O.; Sönnichsen, F.D.; Gelhaus, C.; et al. Characterization and function of the first antibiotic isolated from a vent organism: The extremophile metazoan Alvinella pompejana. PLoS ONE 2014, 9, e95737. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.; Tsarev, A.; Bolosov, I.; Paramonov, A.; Marggraf, M.; Sychev, S.; Shenkarev, Z.; Ovchinnikova, T. Novel antimicrobial peptides from the arctic polychaeta Nicomache minor provide new molecular insight into biological role of the BRICHOS domain. Mar. Drugs 2018, 16, 401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tasiemski, A.; Schikorski, D.; Le Marrec-Croq, F.; Pontoire-Van Camp, C.; Boidin-Wichlacz, C.; Sautière, P.-E. Hedistin: A novel antimicrobial peptide containing bromotryptophan constitutively expressed in the NK cells-like of the marine annelid, Nereis diversicolor. Dev. Comp. Immunol. 2007, 31, 749–762. [Google Scholar] [CrossRef]
- Joo, M.-S.; Choi, K.-M.; Cho, D.-H.; Choi, H.-S.; Min, E.Y.; Han, H.-J.; Cho, M.Y.; Bae, J.-S.; Park, C.-I. The molecular characterization, expression analysis and antimicrobial activity of theromacin from Asian polychaeta (Perinereis linea). Dev. Comp. Immunol. 2020, 112, 103773. [Google Scholar] [CrossRef]
- Seaver, E.C. Annelid models I: Capitella teleta. Curr. Opin. Genet. Dev. 2016, 39, 35–41. [Google Scholar] [CrossRef]
- De Jong, D.M.; Seaver, E.C. Investigation into the cellular origins of posterior regeneration in the annelid Capitella teleta. Regeneration 2018, 5, 61–77. [Google Scholar] [CrossRef]
- Nakamura, T.; Furunaka, H.; Miyata, T.; Tokunaga, F.; Muta, T.; Iwanaga, S.; Niwa, M.; Takao, T.; Shimonishi, Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J. Biol. Chem. 1988, 263, 16709–16713. [Google Scholar]
- Leppert, A.; Chen, G.; Johansson, J. BRICHOS: A chaperone with different activities depending on quaternary structure and cellular location? Amyloid 2019, 26, 152–153. [Google Scholar] [CrossRef]
- Panteleev, P.V.; Balandin, S.V.; Ivanov, V.T.; Ovchinnikova, T.V. A therapeutic potential of animal β-hairpin antimicrobial peptides. Curr. Med. Chem. 2017, 24. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Balandin, S.V.; Ovchinnikova, T.V. Effect of arenicins and other β-hairpin antimicrobial peptides on Pseudomonas aeruginosa PAO1 biofilms. Pharm. Chem. J. 2017, 50, 715–720. [Google Scholar] [CrossRef]
- Pyrkov, T.V.; Chugunov, A.O.; Krylov, N.A.; Nolde, D.E.; Efremov, R.G. PLATINUM: A web tool for analysis of hydrophobic/hydrophilic organization of biomolecular complexes. Bioinformatics 2009, 25, 1201–1202. [Google Scholar] [CrossRef] [PubMed]
- Stavrakoudis, A.; Tsoulos, I.G.; Shenkarev, Z.O.; Ovchinnikova, T.V. Molecular dynamics simulation of antimicrobial peptide arenicin-2: β-Hairpin stabilization by noncovalent interactions. Biopolymers 2009, 92, 143–155. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Balandin, S.V.; Trunov, K.I.; Paramonov, A.S.; Sukhanov, S.V.; Barsukov, L.I.; Arseniev, A.S.; Ovchinnikova, T.V. Molecular mechanism of action of β-hairpin antimicrobial peptide arenicin: Oligomeric structure in dodecylphosphocholine micelles and pore formation in planar lipid bilayers. Biochemistry 2011, 50, 6255–6265. [Google Scholar] [CrossRef]
- Mani, R.; Cady, S.D.; Tang, M.; Waring, A.J.; Lehrer, R.I.; Hong, M. Membrane-dependent oligomeric structure and pore formation of a beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. USA 2006, 103, 16242–16247. [Google Scholar] [CrossRef] [Green Version]
- Panteleev, P.V.; Myshkin, M.Y.; Shenkarev, Z.O.; Ovchinnikova, T.V. Dimerization of the antimicrobial peptide arenicin plays a key role in the cytotoxicity but not in the antibacterial activity. Biochem. Biophys. Res. Commun. 2017, 482, 1320–1326. [Google Scholar] [CrossRef]
- Perczel, A.; Hollosi, M.; Foxman, B.M.; Fasman, G.D. Conformational analysis of pseudocyclic hexapeptides based on quantitative circular dichroism (CD), NOE, and X-ray data. The pure CD spectra of type I and type II.beta.-turns. J. Am. Chem. Soc. 1991, 113, 9772–9784. [Google Scholar] [CrossRef]
- Domingues, T.M.; Perez, K.R.; Miranda, A.; Riske, K.A. Comparative study of the mechanism of action of the antimicrobial peptide gomesin and its linear analogue: The role of the β-hairpin structure. Biochim. Biophys. Acta BBA Biomembr. 2015, 1848, 2414–2421. [Google Scholar] [CrossRef] [Green Version]
- Deplazes, E.; Chin, Y.K.-Y.; King, G.F.; Mancera, R.L. The unusual conformation of cross-strand disulfide bonds is critical to the stability of β-hairpin peptides. Proteins Struct. Funct. Bioinform. 2020, 88, 485–502. [Google Scholar] [CrossRef]
- Sychev, S.V.; Panteleev, P.V.; Ovchinnikova, T.V. Structural study of the β-hairpin marine antimicrobial peptide arenicin-2 in PC/PG lipid bilayers by fourier transform infrared spectroscopy. Russ. J. Bioorganic Chem. 2017, 43, 502–508. [Google Scholar] [CrossRef]
- Sychev, S.V.; Sukhanov, S.V.; Panteleev, P.V.; Shenkarev, Z.O.; Ovchinnikova, T.V. Marine antimicrobial peptide arenicin adopts a monomeric twisted β-hairpin structure and forms low conductivity pores in zwitterionic lipid bilayers. Pept. Sci. 2018, 110, e23093. [Google Scholar] [CrossRef] [PubMed]
- Petrovskaya, L.E.; Lukashev, E.P.; Chupin, V.V.; Sychev, S.V.; Lyukmanova, E.N.; Kryukova, E.A.; Ziganshin, R.H.; Spirina, E.V.; Rivkina, E.M.; Khatypov, R.A.; et al. Predicted bacteriorhodopsin from Exiguobacterium sibiricum is a functional proton pump. FEBS Lett. 2010, 584, 4193–4196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galliard, T. Phospholipid metabolism in photosynthetic plants. In Form and Function of Phospholipids, 2nd ed.; Ansell, G.B., Hawthorne, J.N., Dawson, R.M.C., Eds.; Elsevier: New York, NY, USA, 1973; pp. 253–288. [Google Scholar]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.W.; Wong, G.C.L. Antimicrobial peptides and induced membrane curvature: Geometry, coordination chemistry, and molecular engineering. Curr. Opin. Solid State Mater. Sci. 2013, 17, 151–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savini, F.; Bobone, S.; Roversi, D.; Mangoni, M.L.; Stella, L. From liposomes to cells: Filling the gap between physicochemical and microbiological studies of the activity and selectivity of host-defense peptides. Pept. Sci. 2018, 110, e24041. [Google Scholar] [CrossRef]
- Doherty, T.; Waring, A.J.; Hong, M. Peptide–lipid interactions of the β-hairpin antimicrobial peptide tachyplesin and its linear derivatives from solid-state NMR. Biochim. Biophys. Acta BBA Biomembr. 2006, 1758, 1285–1291. [Google Scholar] [CrossRef] [Green Version]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta BBA Biomembr. 2005, 1717, 67–88. [Google Scholar] [CrossRef] [Green Version]
- Méndez-Vilas, A. Science Against Microbial Pathogens: Communicating Current Research and Technological Advances; Formatex Research Center: Badajoz, Spain, 2011; ISBN 978-84-939843-1-1. [Google Scholar]
- Matsuzaki, K.; Yoneyama, S.; Fujii, N.; Miyajima, K.; Yamada, K.; Kirino, Y.; Anzai, K. Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry 1997, 36, 9799–9806. [Google Scholar] [CrossRef]
- Doherty, T.; Waring, A.J.; Hong, M. Dynamic structure of disulfide-removed linear analogs of tachyplesin-I in the lipid Bilayer from solid-state NMR. Biochemistry 2008, 47, 1105–1116. [Google Scholar] [CrossRef]
- Su, Y.; Li, S.; Hong, M. Cationic membrane peptides: Atomic-level insight of structure–activity relationships from solid-state NMR. Amino Acids 2013, 44, 821–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panteleev, P.V.; Bolosov, I.A.; Kalashnikov, A.À.; Kokryakov, V.N.; Shamova, O.V.; Emelianova, A.A.; Balandin, S.V.; Ovchinnikova, T.V. Combined antibacterial effects of goat cathelicidins with different mechanisms of action. Front. Microbiol. 2018, 9, 2983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciudad, S.; Puig, E.; Botzanowski, T.; Meigooni, M.; Arango, A.S.; Do, J.; Mayzel, M.; Bayoumi, M.; Chaignepain, S.; Maglia, G.; et al. Aβ(1-42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat. Commun. 2020, 11, 3014. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, A.; Kuwahara, J.; Fujii, N.; Sugiura, Y. Binding of tachyplesin I to DNA revealed by footprinting analysis: Significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 1992, 31, 2998–3004. [Google Scholar] [CrossRef] [PubMed]
- Elliott, A.G.; Huang, J.X.; Neve, S.; Zuegg, J.; Edwards, I.A.; Cain, A.K.; Boinett, C.J.; Barquist, L.; Lundberg, C.V.; Steen, J.; et al. An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria. Nat. Commun. 2020, 11, 3184. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Qi, J.; Shan, B.; Ma, Y. Tachyplesin causes membrane instability that kills multidrug-resistant bacteria by inhibiting the 3-ketoacyl carrier protein reductase FabG. Front. Microbiol. 2018, 9, 825. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Ovchinnikova, T.V. Improved strategy for recombinant production and purification of antimicrobial peptide tachyplesin I and its analogs with high cell selectivity. Biotechnol. Appl. Biochem. 2017, 64, 35–42. [Google Scholar] [CrossRef]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Blaskovich, M.A.T.; Cooper, M.A. Structure–activity and −toxicity relationships of the antimicrobial peptide tachyplesin-1. ACS Infect. Dis. 2017, 3, 917–926. [Google Scholar] [CrossRef]
- Kushibiki, T.; Kamiya, M.; Aizawa, T.; Kumaki, Y.; Kikukawa, T.; Mizuguchi, M.; Demura, M.; Kawabata, S.; Kawano, K. Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochim. Biophys. Acta BBA Proteins Proteom. 2014, 1844, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, E.; Güntert, P. Automated structure determination from NMR spectra. In Structural Proteomics; Owens, R.J., Ed.; Springer: New York, NY, USA, 2015; Volume 1261, pp. 303–329. ISBN 978-1-4939-2229-1. [Google Scholar]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef] [Green Version]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Genet. 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Nadezhdin, K.D.; Sobol, V.A.; Sobol, A.G.; Skjeldal, L.; Arseniev, A.S. Conformation and mode of membrane interaction in cyclotides. FEBS J. 2006, 273, 2658–2672. [Google Scholar] [CrossRef]
- Huang, K.S.; Bayley, H.; Liao, M.J.; London, E.; Khorana, H.G. Refolding of an integral membrane protein. Denaturation, renaturation, and reconstitution of intact bacteriorhodopsin and two proteolytic fragments. J. Biol. Chem. 1981, 256, 3802–3809. [Google Scholar] [PubMed]
- Kuzmin, D.V.; Emelianova, A.A.; Kalashnikova, M.B.; Panteleev, P.V.; Balandin, S.V.; Serebrovskaya, E.O.; Belogurova-Ovchinnikova, O.Y.; Ovchinnikova, T.V. Comparative in vitro study on cytotoxicity of recombinant β-hairpin peptides. Chem. Biol. Drug Des. 2018, 91, 294–303. [Google Scholar] [CrossRef]








| Peptide | Recombinant Peptide Final Yield, mg/L | RP-HPLC Retention Time, min | Hydrophobicity Index 1/Hydrophobic Residues, % 2 | Calculated [M + H]+ Monoisotopic Molecular Mass, Da 3 | Measured Monoisotopic m/z Value 4 |
|---|---|---|---|---|---|
| Capitellacin | 6.1 | 38.5 | −0.215/45 | 2379.16 | 2379.30 |
| Tachyplesin-1 | 7.2 | 37.5 | −0.482/47 | 2264.10 | 2263.73 |
| Environment | Dielectric Constant | λmax (nm) | Ksv (M−1) |
|---|---|---|---|
| H2O | 80.4 | 350 | 9.9 |
| CH3OH | 33.6 | 335 | - |
| PE/PG | ~2 * | 332 | 3.1 |
| Bacteria | Minimum Inhibitory Concentration (µM) | ||
|---|---|---|---|
| Capitellacin | Tachyplesin-1 | Polymyxin B | |
| Gram-positive | |||
| S. aureus ATCC 6538P | 8 | 1 | n.d. |
| M. luteus B-1314 | 4 | 2 | n.d. |
| B. subtilis B-886 | 16 | 2 | n.d. |
| Gram-negative | |||
| E. coli ATTC 25922 | 0.5 | 0.25 | 0.06 |
| E. coli ML-35p | 0.25 | 0.125 | 0.06 |
| E. coli CI 214 | 0.125 | 0.06 | 0.03 |
| E. coli (MDR CI 1057) | 0.25 | 0.125 | 0.06 |
| E. coli (MDR CI 3600) | 0.5 | 0.06 | 0.06 |
| E. cloacae (XDR CI 4172) | 4 | 0.25 | 0.5 |
| A. baumanii (XDR CI 2675) | 0.25 | 0.016 | 0.5 |
| A. baumanii (XDR CI 450) | 0.5 | 0.06 | 0.03 |
| K. pneumonia ATCC 700603) | 4 | 1 | 0.125 |
| K. pneumonia (MDR CI 358) | 2 | 1 | 0.06 |
| K. pneumonia (XDR CI 1056) | 4 | 0.5 | 0.5 |
| K. pneumonia (XDR CI 3395) | 2 | 0.5 | 0.125 |
| P. aeruginosa ATCC 27853 | 2 | 0.25 | 0.125 |
| P. aeruginosa PAO1 | 2 | 0.25 | 0.25 |
| P. aeruginosa (MDR CI 223 *) | 8 | 1 | 0.5 |
| P. aeruginosa (XDR CI 236 *) | 4 | 0.5 | 0.5 |
| P. aeruginosa (XDR CI 1049 *) | 4 | 1 | 0.25 |
| P. aeruginosa (XDR CI 1995 *) | 8 | 1 | 0.125 |
| S. marcescens (MDR CI 1689) | >32 | >32 | >32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panteleev, P.V.; Tsarev, A.V.; Safronova, V.N.; Reznikova, O.V.; Bolosov, I.A.; Sychev, S.V.; Shenkarev, Z.O.; Ovchinnikova, T.V. Structure Elucidation and Functional Studies of a Novel β-hairpin Antimicrobial Peptide from the Marine Polychaeta Capitella teleta. Mar. Drugs 2020, 18, 620. https://doi.org/10.3390/md18120620
Panteleev PV, Tsarev AV, Safronova VN, Reznikova OV, Bolosov IA, Sychev SV, Shenkarev ZO, Ovchinnikova TV. Structure Elucidation and Functional Studies of a Novel β-hairpin Antimicrobial Peptide from the Marine Polychaeta Capitella teleta. Marine Drugs. 2020; 18(12):620. https://doi.org/10.3390/md18120620
Chicago/Turabian StylePanteleev, Pavel V., Andrey V. Tsarev, Victoria N. Safronova, Olesia V. Reznikova, Ilia A. Bolosov, Sergei V. Sychev, Zakhar O. Shenkarev, and Tatiana V. Ovchinnikova. 2020. "Structure Elucidation and Functional Studies of a Novel β-hairpin Antimicrobial Peptide from the Marine Polychaeta Capitella teleta" Marine Drugs 18, no. 12: 620. https://doi.org/10.3390/md18120620
APA StylePanteleev, P. V., Tsarev, A. V., Safronova, V. N., Reznikova, O. V., Bolosov, I. A., Sychev, S. V., Shenkarev, Z. O., & Ovchinnikova, T. V. (2020). Structure Elucidation and Functional Studies of a Novel β-hairpin Antimicrobial Peptide from the Marine Polychaeta Capitella teleta. Marine Drugs, 18(12), 620. https://doi.org/10.3390/md18120620