Cytotoxic Thiodiketopiperazine Derivatives from the Deep Sea-Derived Fungus Epicoccum nigrum SD-388
Abstract
:1. Introduction
2. Results and Discussion
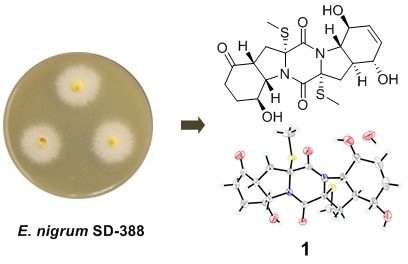
2.1. Structure Elucidation of the New Compounds
2.2. Biological Activities of the Isolated Compounds
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation
3.4. Extraction and Isolation
3.5. X-Ray Crystallographic Analysis
3.6. Computational Section
3.7. Cytotoxic Assays
3.7.1. Cell Culture
3.7.2. Cell Proliferation Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Niu, S.; Xia, M.; Chen, M.; Liu, X.; Li, Z.; Xie, Y.; Shao, Z.; Zhang, G. Cytotoxic polyketides isolated from the deep-sea-derived fungus Penicillium chrysogenum MCCC 3A00292. Mar. Drugs 2019, 17, 686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Fan, Z.; Sun, Z.; Liu, H.; Zhang, W. Dechdigliotoxins A–C, three novel disulfide-bridged gliotoxin dimers from deep-sea sediment derived fungus Dichotomomyces cejpii. Mar. Drugs 2019, 17, 596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, S.; Che, Y.; Liu, X. Epicoccins A–D, epipolythiodioxopiperazines from a Cordyceps-colonizing isolate of Epicoccum nigrum. J. Nat. Prod. 2007, 70, 1522–1525. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Sun, B.; Gao, H.; Chen, X.; Liu, S.; Yao, X.; Liu, X.; Che, Y. Diketopiperazines from the Cordyceps-colonizing fungus Epicoccum nigrum. J. Nat. Prod. 2009, 72, 2115–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Z.; Wen, S.; Ding, M.; Guo, H.; Huang, C.; Zhu, X.; Huang, J.; She, Z.; Long, Y. The purification, characterization, and biological activity of new polyketides from mangrove-derived endophytic fungus Epicoccum nigrum SCNU-F0002. Mar. Drugs 2019, 17, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.H.; Mao, W.J.; Jiao, J.Y.; Xu, J.C.; Li, H.Y.; Chen, Y.; Xu, J.; Zhao, C.Q.; Hou, Y.J.; Yang, Y.P. Structural characterization of extracellular polysaccharides produced by the marine fungus Epicoccum nigrum JJY-40 and their antioxidant activities. Mar. Biotechnol. 2011, 13, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.X.; Jensen, P.R.; Williams, P.G.; Fenical, W. Isolation and structure assignments of rostratins A−D, cytotoxic disulfides produced by the marine-derived fungus Exserohilum rostratum. J. Nat. Prod. 2004, 67, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.H.; Li, X.M.; Lv, C.T.; Huang, C.G.; Wang, B.G. Brocazines A–F, cytotoxic bisthiodiketopiperazine derivatives from Penicillium brocae MA-231, an endophytic fungus derived from the marine mangrove plant Avicennia marina. J. Nat. Prod. 2014, 77, 1921–1927. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.M.; Meng, L.H.; Jiang, W.L.; Xu, G.M.; Huang, C.G.; Wang, B.G. Bisthiodiketopiperazines and acorane sesquiterpenes produced by the marine-derived fungus Penicillium adametzioides AS-53 on different culture media. J. Nat. Prod. 2015, 78, 1294–1299. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Li, X.M.; Meng, L.H.; Konuklugil, B.; Li, X.; Li, H.L.; Wang, B.G. Isolation and characterization of three pairs of indolediketopiperazine enantiomers containing infrequent N-methoxy substitution from the marine algal-derived endophytic fungus Acrostalagmus luteoalbus TK-43. Bioorg. Chem. 2019, 90, 103030. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Li, X.M.; Li, X.; Xu, G.M.; Liu, Y.; Wang, B.G. Aspewentins D–H, 20-nor-isopimarane derivatives from the deep sea sediment-derived fungus Aspergillus wentii SD-310. J. Nat. Prod. 2016, 79, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chi, L.P.; Navarro-Vázquez, A.; Hwang, S.; Schmieder, P.; Li, X.M.; Li, X.; Yang, S.Q.; Lei, X.; Wang, B.G.; et al. Stereochemical elucidation of natural products from residual chemical shift anisotropies in a liquid crystalline phase. J. Am. Chem. Soc. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawahara, N.; Nozawa, K.; Nakajima, S.; Kawai, K.I. Emeheterone, a pyrazinone derivative from Emericella heterothallica. Phytochemistry 1988, 27, 3022–3024. [Google Scholar] [CrossRef]
- Wang, J.M.; Jiang, N.; Ma, J.; Yu, S.S.; Tan, R.X.; Dai, J.G.; Si, Y.K.; Ding, G.Z.; Ma, S.G.; Qu, L.; et al. Study on absolute configurations of α/α′ chiral carbons of thiodiketopiperazines by experimental and calculated circular dichroism spectra. Tetrahedron 2013, 69, 1195–1201. [Google Scholar] [CrossRef]
- Crystallographic data of compounds 1–3 have been deposited in the Cambridge Crystallographic Data Centre as CCDC 1935572, CCDC 1935573 and CCDC 1935571, respectively. Available online: https://www.ccdc.cam.ac.uk/structures/? (accessed on 13 March 2020).
- Sheldrick, G.M. SADABS, Software for empirical absorption correction; University of Gottingen: Gottingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. SHELXTL, Structure determination software programs; Bruker Analytical X-ray System Inc.: Madison, WI, USA, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXL-97 and SHELXS-97, Program for X-ray Crystal Structure Solution and Refinement; University of Gottingen: Gottingen, Germany, 1997. [Google Scholar]
- Flack, H.D. On enantiomorph-polarity estimation. Acta Cryst. 1983, 39, 876–881. [Google Scholar] [CrossRef]
- Gaussian09, RevisionC.01. Available online: http://gaussian.com/g09citation/ (accessed on 13 March 2020).
- Liu, G.; Kuang, S.; Cao, R.; Wang, J.; Peng, Q.; Sun, C. Sorafenib kills liver cancer cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids via the ATP-AMPK-mTOR-SREBP1 signaling pathway. FASEB J. 2019, 33, 10089–10103. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| No. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 2 | 4.17, t (6.4) | ||||
| 3 | α 2.80, d (13.4) | α 2.75, d (13.5) | α 3.13, d (17.7) | α 2.68, m | a 3.02, dd (13.3, 6.4) |
| β 2.28, m | β 2.35, dd (13.5, 8.5) | β 2.89, m | β 2.99, dd (14.8, 7.9) | b 2.95, dd (13.3, 6.4) | |
| 4 | 2.98, t (8.0) | 2.92, t (8.5) | 3.06, t (8.3) | 3.18, t (7.9) | |
| 6 | α 2.22, m | α 2.54, dd (17.1, 4.7) | α 2.85, m | α 2.71, m | 6.79, d (7.3) |
| β 2.61, m | β 2.65, m | β 2.92, m | β 2.61, m | ||
| 7 | α 2.19, m | 3.82, ddd (10.1, 4.7, 2.0) | 3.73, m | 3.29, ddd (10.9, 4.7, 1.5) | 6.96, m |
| β 1.92, m | |||||
| 8 | 4.36, m | 4.56, m | 4.01, dt (4.6, 2.4) | 5.03, t (4.7) | 6.67, td (7.3, 1.2) |
| 9 | 4.33, m | 4.33, m | 4.67, dd (8.3, 2.4) | 4.41, dd (7.9, 4.7) | 6.98, m |
| 3’ | α 2.42, dd (12.5,4.9) | α 2.73, d (13.4) | α 2.93, m | α 2.74, m | 6.83, s |
| β 2.26, m | β 2.34, dd (13.4, 8.5) | β 3.29, d (19.4) | β 2.93, dd (14.8, 8.0) | ||
| 4’ | 2.09, m | 2.91, t (8.5) | 3.17, d (8.1) | 3.23, t (8.0) | |
| 5’ | 4.11, m | 7.27, d (7.3) | |||
| 6’ | 5.68, d (9.8) | α 2.40, dd (16.8, 4.4) | α 2.89, m | α 2.66, m | 7.40, t (7.3) |
| β 2.61, m | β 2.97, m | β 2.27, dt (16.7, 4.1) | |||
| 7’ | 5.53, d (9.8) | 4.12, ddd (10.2, 4.4, 2.0) | 3.92, m | α 1.62, td (12.5, 5.0) | 7.32, t (7.3) |
| β 1.89, m | |||||
| 8’ | 4.08, m | 4.29, s | 5.04, dd (4.3, 2.0) | 4.81, m | 7.40, t (7.3) |
| 9’ | 3.41, dd (12.0, 8.1) | 4.34, m | 4.83, dd (8.1, 2.0) | 4.36, m | 7.27, d (7.3) |
| 1-NMe | 2.69, s | ||||
| 1’-NH | -b | ||||
| 2-SMe | 1.95, s | 1.93, s | |||
| 2’-SMe | 2.08, s | 1.92, s | |||
| 2″ | 2.04, s | ||||
| 5-OH | 8.50, br s | ||||
| 8-OH | 5.36, d (3.2) | 5.44, br s | 6.25, d (2.4) | 5.64, d (4.7) | |
| 5’-OH | 5.92, s | ||||
| 7’-OH | 5.19, br s | ||||
| 8’-OH | 5.28, d (5.8) | 5.44, br s | 5.49, d (4.0) | ||
| 7-OMe | 3.25, s | 3.21, s |
| No. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 168.7, C | 165.5, C | 158.3, C | 162.2, C | 167.2, C |
| 2 | 71.4, C | 71.6, C | 71.1, C | 76.2, C | 55.6, CH |
| 3 | 34.3, CH2 | 34.4, CH2 | 41.4, CH2 | 32.6, CH2 | 34.8, CH2 |
| 4 | 44.0, CH | 43.8, CH | 45.0, CH | 46.4, CH | 122.0, C |
| 5 | 207.5, C | 206.6, C | 207.1, C | 207.8, C | 156.1, C |
| 6 | 33.8, CH2 | 40.4, CH2 | 43.2, CH2 | 40.7, CH2 | 115.1, CH |
| 7 | 25.9, CH2 | 75.8, CH | 41.3, CH | 75.5, CH | 127.8, CH |
| 8 | 63.6, CH | 61.5, CH b | 65.2, CH | 61.9, CH | 118.4, CH |
| 9 | 64.8, CH | 63.2, CH | 60.3, CH | 63.2, CH | 131.2, CH |
| 1’ | 165.5, C | 165.4, C | 158.9, C | 162.2, C | 162.4, C |
| 2’ | 72.9, C | 71.6, C | 71.7, C | 76.4, C | 132.2, C |
| 3’ | 35.1, CH2 | 34.1, CH2 | 41.5, CH2 | 32.2, CH2 | 117.7, CH |
| 4’ | 43.4, CH | 43.5, CH | 45.3, CH | 46.7, CH | 133.8, C |
| 5’ | 71.3, CH | 207.4, C | 206.3, C | 208.6, C | 129.4, CH |
| 6’ | 133.3, CH | 43.3, CH2 | 43.5, CH2 | 33.9, CH2 | 128.1, CH |
| 7’ | 129.9, CH | 65.7, CH | 38.8, CH | 25.4, CH2 | 127.9, CH |
| 8’ | 68.9, CH | 68.1, CH b | 67.4, CH | 60.8, CH | 128.1, CH |
| 9’ | 67.8, CH | 62.7, CH | 57.5, CH | 65.8, CH | 129.4, CH |
| 1″ | 168.8, C | ||||
| 2″ | 20.6, CH3 | ||||
| 1-NMe | 34.5, CH3 | ||||
| 2-SMe | 14.4, CH3 | 14.2, CH3 | |||
| 2’-SMe | 14.2, CH3 | 13.9, CH3 | |||
| 7-OMe | 55.8, CH3 | 56.0, CH3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, L.-P.; Li, X.-M.; Li, L.; Li, X.; Wang, B.-G. Cytotoxic Thiodiketopiperazine Derivatives from the Deep Sea-Derived Fungus Epicoccum nigrum SD-388. Mar. Drugs 2020, 18, 160. https://doi.org/10.3390/md18030160
Chi L-P, Li X-M, Li L, Li X, Wang B-G. Cytotoxic Thiodiketopiperazine Derivatives from the Deep Sea-Derived Fungus Epicoccum nigrum SD-388. Marine Drugs. 2020; 18(3):160. https://doi.org/10.3390/md18030160
Chicago/Turabian StyleChi, Lu-Ping, Xiao-Ming Li, Li Li, Xin Li, and Bin-Gui Wang. 2020. "Cytotoxic Thiodiketopiperazine Derivatives from the Deep Sea-Derived Fungus Epicoccum nigrum SD-388" Marine Drugs 18, no. 3: 160. https://doi.org/10.3390/md18030160
APA StyleChi, L. -P., Li, X. -M., Li, L., Li, X., & Wang, B. -G. (2020). Cytotoxic Thiodiketopiperazine Derivatives from the Deep Sea-Derived Fungus Epicoccum nigrum SD-388. Marine Drugs, 18(3), 160. https://doi.org/10.3390/md18030160