Insights into the Light Response of Skeletonema marinoi: Involvement of Ovothiol
Abstract
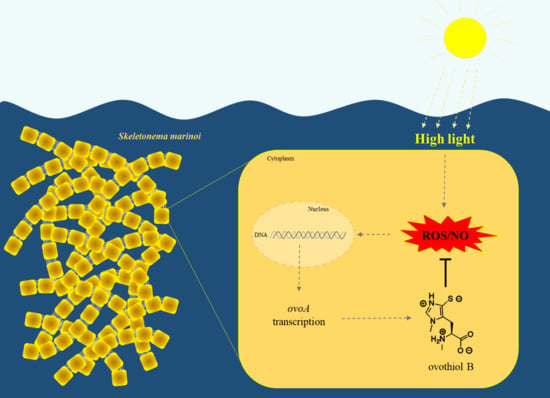
:1. Introduction
2. Results
2.1. Molecular Response to High Light Conditions
2.2. Molecular Response to Prolonged Darkness and Low Light Conditions
3. Discussion
4. Materials and Methods
4.1. Experimental Strategy and Sampling
- −
- Darkness (continuous absence of light; 0 h:24 h light:dark; daily light dose: 0 mol photons m−2);
- −
- Very low sinusoidal light 10 (midday light intensity peak: 10 µmol photons s−1 m−2; 12 h:12 h light:dark; daily light dose: 0.24 mol photons m−2; Sin10);
- −
- Very low square-wave light 10 (continuous light intensity: 10 µmol photons s−1 m−2; 24 h:0 h light:dark; daily light dose: 1 mol photons m−2; Square10);
- −
- Low sinusoidal light 150 (midday light intensity peak: 150 µmol photons s−1 m−2; 12 h:12 h light:dark; daily light dose: 3.6 mol photons m−2; Sin150);
- −
- High sinusoidal light 600 (midday light intensity peak: 600 µmol photons s−1 m−2; 12 h:12 h light:dark; daily light dose: 14.4 mol photons m−2; Sin600);
- −
- High square-wave light 300 (continuous light intensity: 300 µmol photons s−1 m−2; 12 h:12 h light:dark; daily light dose: 14.4 mol photons m−2; Square300);
- −
- High square-wave light 600 (continuous light intensity: 600 µmol photons s−1 m−2; 12 h:12 h light:dark; daily light dose: 28.8 mol photons m−2; Square600).
4.2. Cell Density
4.3. RNA Extraction, Reverse Transcription and Best Reference Genes Assessment
4.4. Reverse Transcription-Quantitative PCR (RT-qPCR) Experiments
4.5. Nitric Oxide (NO) Determination
4.6. Reactive Oxygen Species (ROS) Determination
4.7. Thiols Determination
4.8. Data Analysis
4.9. Nitric Oxide Synthase (Nos) Protein Sequence Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nelson, D.M.; Treguer, P.; Brzezinski, M.A.; Leynaert, A.; Queguiner, B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycles 1995, 9, 359–372. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Raven, J.A. Aquatic Photosynthesis, 2nd ed.; Princeton University Press: Princeton, NJ, USA, 2007. [Google Scholar]
- Barra, L.; Chandrasekaran, R.; Corato, F.; Brunet, C. The Challenge of Ecophysiological Biodiversity for Biotechnological Applications of Marine Microalgae. Mar. Drugs 2014, 12, 1641–1675. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Lu, Y.; Zheng, J.W.; Yang, W.D.; Liu, J.S. Biochemical and genetic engineering of diatoms for polyunsaturated fatty acid biosynthesis. Mar. Drugs 2014, 12, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Dolch, L.J.; Marechal, E. Inventory of fatty acid desaturases in the pennate diatom Phaeodactylum tricornutum. Mar. Drugs 2015, 13, 1317–1339. [Google Scholar] [CrossRef] [Green Version]
- Baldisserotto, C.; Sabia, A.; Ferroni, L.; Pancaldi, S. Biological aspects and biotechnological potential of marine diatoms in relation to different light regimens. World J. Microbiol. Biotechnol. 2019, 35, 35. [Google Scholar] [CrossRef]
- Galasso, C.; Gentile, A.; Orefice, I.; Ianora, A.; Bruno, A.; Noonan, D.M.; Sansone, C.; Albini, A.; Brunet, C. Microalgal Derivatives as Potential Nutraceutical and Food Supplements for Human Health: A Focus on Cancer Prevention and Interception. Nutrients 2019, 11, 1226. [Google Scholar] [CrossRef] [Green Version]
- Montsant, A.; Allen, A.E.; Coesel, S.; Martino, A.D.; Falciatore, A.; Mangogna, M.; Siaut, M.; Heijde, M.; Jabbari, K.; Maheswari, U.; et al. Identification and comparative genomic analysis of signaling and regulatory components in the diatom Thalassiosira pseudonana. J. Phycol. 2007, 43, 585–604. [Google Scholar] [CrossRef]
- Parker, M.S.; Mock, T.; Armbrust, E.V. Genomic Insights into Marine Microalgae. Annu. Rev. Genet. 2008, 42, 619–645. [Google Scholar] [CrossRef]
- Kooistra, W.H.; Gersonde, R.; Medlin, L.K.; Mann, D.G. The Origin and Evolution of the Diatoms: Their Adaptation to a Planktonic Existence. In Evolution of Primary Producers in the Sea; Falkowski, P.G., Knoll, A.H., Eds.; Academic Press: Burlington, MA, USA, 2007; pp. 207–249. [Google Scholar]
- Vardi, A. Cell signaling in marine diatoms. Commun. Integr. Biol. 2008, 1, 134–136. [Google Scholar] [CrossRef] [Green Version]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Armbrust, E.V. The life of diatoms in the world’s oceans. Nature 2009, 459, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Brunet, C.; Chandrasekaran, R.; Barra, L.; Giovagnetti, V.; Corato, F.; Ruban, A.V. Spectral radiation dependent photoprotective mechanism in the diatom Pseudo-nitzschia multistriata. PLoS ONE 2004, 9, e87015. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, M. Carotenoid biosynthesis in diatoms. Photosynth. Res. 2010, 106, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, R.; Barra, L.; Carillo, S.; Caruso, T.; Corsaro, M.M.; Dal Piaz, F.; Graziani, G.; Corato, F.; Pepe, D.; Manfredonia, A.; et al. Light modulation of biomass and macromolecular composition of the diatom Skeletonema marinoi. J. Biotechnol. 2014, 192, 114–122. [Google Scholar] [CrossRef]
- Orefice, I.; Chandrasekaran, R.; Smerilli, A.; Corato, F.; Caruso, T.; Casillo, A.; Corsaro, M.M.; Dal Piaz, F.; Ruban, A.V.; Brunet, C. Light-induced changes in the photosynthetic physiology and biochemistry in the diatom Skeletonema marinoi. Algal Res. 2016, 17, 1–13. [Google Scholar] [CrossRef]
- Smerilli, A.; Orefice, I.; Corato, F.; Gavalas Olea, A.; Ruban, A.V.; Brunet, C. Photoprotective and antioxidant responses to light spectrum and intensity variations in the coastal diatom Skeletonema marinoi. Environ. Microbiol. 2017, 19, 611–627. [Google Scholar] [CrossRef]
- Smerilli, A.; Balzano, S.; Maselli, M.; Blasio, M.; Orefice, I.; Galasso, C.; Sansone, C.; Brunet, C. Antioxidant and Photoprotection Networking in the Coastal Diatom Skeletonema marinoi. Antioxidants 2019, 8, 154. [Google Scholar] [CrossRef] [Green Version]
- Kirk, J.T.O. Thermal dissociation of fucoxanthin-protein binding in pigment complexes from chloroplasts of Hormosira (phaeophyta). Plant Sci. Lett. 1977, 9, 373–380. [Google Scholar] [CrossRef]
- Müller, P.; Li, X.-P.; Niyogi, K.K. Non-Photochemical Quenching. A Response to Excess Light Energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef] [Green Version]
- Lavaud, J. Fast Regulation of Photosynthesis in Diatoms: Mechanisms, Evolution and Ecophysiology. Funct. Plant Sci. Biotechnol. 2007, 1, 267–287. [Google Scholar]
- Milito, A.; Castellano, I.; Burn, R.; Seebeck, F.P.; Brunet, C.; Palumbo, A. First evidence of ovothiol biosynthesis in marine diatoms. Free Radic. Biol. Med. 2020, 152, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Turner, E.; Klevit, R.; Hager, L.J.; Shapiro, B.M. Ovothiols, a family of redox-active mercaptohistidine compounds from marine invertebrate eggs. Biochemistry 1987, 26, 4028–4036. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Castellano, I.; Napolitano, A. Ovothiol: A potent natural antioxidant from marine organisms. In Blue Biotechnology. Production and Use of Marine Molecules. Part 2: Marine Molecules for Disease Treatment/Prevention and for Biological Research; La Barre, S., Bates, S.S., Eds.; Wiley VCH: Weinheim, DE, USA, 2018; pp. 583–610. [Google Scholar]
- Weaver, K.H.; Rabenstein, D.L. Thiol/Disulfide Exchange Reactions of Ovothiol A with Glutathione. J. Org. Chem. 1995, 60, 1904–1907. [Google Scholar] [CrossRef]
- Marjanovic, B.; Simic, M.G.; Jovanovic, S.V. Heterocyclic thiols as antioxidants: Why Ovothiol C is a better antioxidant than ergothioneine. Free Radic. Biol. Med. 1995, 18, 679–685. [Google Scholar] [CrossRef]
- Ariyanayagam, M.R.; Fairlamb, A.H. Ovothiol and trypanothione as antioxidants in trypanosomatids. Mol. Biochem. Parasitol. 2001, 115, 189–198. [Google Scholar] [CrossRef]
- Jacob, C. A scent of therapy: Pharmacological implications of natural products containing redox-active sulfur atoms. Nat. Prod. Rep. 2006, 23, 851–863. [Google Scholar] [CrossRef]
- Brancaccio, M.; D’Argenio, G.; Lembo, V.; Palumbo, A.; Castellano, I. Antifibrotic Effect of Marine Ovothiol in an In Vivo Model of Liver Fibrosis. Oxid. Med. Cell Longev. 2018, 2018, 5045734. [Google Scholar] [CrossRef] [Green Version]
- Milito, A.; Brancaccio, M.; D’Argenio, G.; Castellano, I. Natural Sulfur-Containing Compounds: An Alternative Therapeutic Strategy against Liver Fibrosis. Cells 2019, 8, 1356. [Google Scholar] [CrossRef] [Green Version]
- Russo, G.L.; Russo, M.; Castellano, I.; Napolitano, A.; Palumbo, A. Ovothiol isolated from sea urchin oocytes induces autophagy in the Hep-G2 cell line. Mar. Drugs 2014, 12, 4069–4085. [Google Scholar] [CrossRef] [Green Version]
- Brancaccio, M.; Russo, M.; Masullo, M.; Palumbo, A.; Russo, G.L.; Castellano, I. Sulfur-containing histidine compounds inhibit γ-glutamyl transpeptidase activity in human cancer cells. J. Biol. Chem. 2019, 294, 14603–14614. [Google Scholar] [CrossRef]
- Milito, A.; Brancaccio, M.; Lisurek, M.; Masullo, M.; Palumbo, A.; Castellano, I. Probing the Interactions of Sulfur-Containing Histidine Compounds with Human Gamma-Glutamyl Transpeptidase. Mar. Drugs 2019, 17, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellano, I.; Di Tomo, P.; Di Pietro, N.; Mandatori, D.; Pipino, C.; Formoso, G.; Napolitano, A.; Palumbo, A. Anti-Inflammatory Activity of Marine Ovothiol A in an In Vitro Model of Endothelial Dysfunction Induced by Hyperglycemia. Oxid. Med. Cell. Longev. 2018, 2018, 2087373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braunshausen, A.; Seebeck, F.P. Identification and Characterization of the First Ovothiol Biosynthetic Enzyme. J. Am. Chem. Soc. 2011, 133, 1757–1759. [Google Scholar] [CrossRef] [PubMed]
- Naowarojna, N.; Huang, P.; Cai, Y.; Song, H.; Wu, L.; Cheng, R.; Li, Y.; Wang, S.; Lyu, H.; Zhang, L.; et al. In Vitro Reconstitution of the Remaining Steps in Ovothiol A Biosynthesis: C–S Lyase and Methyltransferase Reactions. Org. Lett. 2018, 20, 5427–5430. [Google Scholar] [CrossRef]
- Selman-Reimer, S.; Duhe, R.J.; Stockman, B.J.; Selman, B.R. L-1-N-methyl-4-mercaptohistidine disulfide, a potential endogenous regulator in the redox control of chloroplast coupling factor 1 in Dunaliella. J. Biol. Chem. 1991, 266, 182–188. [Google Scholar]
- O’Neill, E.C.; Trick, M.; Hill, L.; Rejzek, M.; Dusi, R.G.; Hamilton, C.J.; Zimba, P.V.; Henrissat, B.; Field, R.A. The transcriptome of Euglena gracilis reveals unexpected metabolic capabilities for carbohydrate and natural product biochemistry. Mol. Biosyst. 2015, 11, 2808–2820. [Google Scholar] [CrossRef] [Green Version]
- Crawford, N.M.; Guo, F.-Q. New insights into nitric oxide metabolism and regulatory functions. Trends Plant Sci. 2005, 10, 195–200. [Google Scholar] [CrossRef]
- Gechev, T.S.; Van Breusegem, F.; Stone, J.M.; Denev, I.; Laloi, C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 2006, 28, 1091–1101. [Google Scholar] [CrossRef]
- Grün, S.; Lindermayr, C.; Sell, S.; Durner, J. Nitric oxide and gene regulation in plants. J. Exp. Bot. 2006, 57, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Gallina, A.A.; Brunet, C.; Palumbo, A.; Casotti, R. The Effect of Polyunsaturated Aldehydes on Skeletonema marinoi (Bacillariophyceae): The Involvement of Reactive Oxygen Species and Nitric Oxide. Mar. Drugs 2014, 12, 4165–4187. [Google Scholar] [CrossRef] [Green Version]
- Di Dato, V.; Musacchia, F.; Petrosino, G.; Patil, S.; Montresor, M.; Sanges, R.; Ferrante, M.I. Transcriptome sequencing of three Pseudo-nitzschia species reveals comparable gene sets and the presence of Nitric Oxide Synthase genes in diatoms. Sci. Rep. 2015, 5, 12329. [Google Scholar] [CrossRef] [PubMed]
- Barra, L.; Ruggiero, M.V.; Sarno, D.; Montresor, M.; Kooistra, W.H.C.F. Strengths and weaknesses of microarray approaches to detect Pseudo-nitzschia species in the field. Environ. Sci. Pollut. Res. Int. 2013, 20, 6705–6718. [Google Scholar] [CrossRef] [PubMed]
- Andreakis, N.; D’Aniello, S.; Albalat, R.; Patti, F.P.; Garcia-Fernàndez, J.; Procaccini, G.; Sordino, P.; Palumbo, A. Evolution of the Nitric Oxide Synthase Family in Metazoans. Mol. Biol. Evol. 2011, 28, 163–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, B.M. The control of oxidant stress at fertilization. Science 1991, 252, 533–536. [Google Scholar] [CrossRef]
- Castellano, I.; Migliaccio, O.; D’Aniello, S.; Merlino, A.; Napolitano, A.; Palumbo, A. Shedding light on ovothiol biosynthesis in marine metazoans. Sci. Rep. 2016, 6, 21506. [Google Scholar] [CrossRef] [Green Version]
- Yanshole, V.V.; Yanshole, L.V.; Zelentsova, E.A.; Tsentalovich, Y.P. Ovothiol A is the Main Antioxidant in Fish Lens. Metabolites 2019, 9, 95. [Google Scholar] [CrossRef] [Green Version]
- Spies, H.S.; Steenkamp, D.J. Thiols of intracellular pathogens. Identification of ovothiolA in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Europ. J. Biochem. 1994, 224, 203–213. [Google Scholar] [CrossRef]
- Tarrant, A.M.; Payton, S.L.; Reitzel, A.M.; Porter, D.T.; Jenny, M.J. Ultraviolet radiation significantly enhances the molecular response to dispersant and sweet crude oil exposure in Nematostella vectensis. Mar. Environ. Res. 2018, 134, 96–108. [Google Scholar] [CrossRef] [Green Version]
- Diaz de Cerio, O.; Reina, L.; Squatrito, V.; Etxebarria, N.; Gonzalez-Gaya, B.; Cancio, I. Gametogenesis-Related Fluctuations in Ovothiol Levels in the Mantle of Mussels from Different Estuaries: Fighting Oxidative Stress for Spawning in Polluted Waters. Biomolecules 2020, 10, 373. [Google Scholar] [CrossRef] [Green Version]
- Rohl, I.; Schneider, B.; Schmidt, B.; Zeeck, E. L-ovothiol A: The egg release pheromone of the marine polychaete Platynereis Dumerilii: Anellida: Polychaeta. Z. Naturforsch. C J. Biosci. 1999, 54, 1145–1147. [Google Scholar] [CrossRef]
- Castellano, I.; Seebeck, F.P. On ovothiol biosynthesis and biological roles: From life in the ocean to therapeutic potential. Nat. Prod. Rep. 2018, 35, 1241–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerdol, M.; Sollitto, M.; Pallavicini, A.; Castellano, I. The complex evolutionary history of sulfoxide synthase in ovothiol biosynthesis. Proc. R. Soc. B 2019, 286, 20191812. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, F.; Martin, A.; McMinn, A. Insights into the Production and Role of Nitric Oxide in the Antarctic Sea-ice Diatom Fragilariopsis cylindrus. J. Phycol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Sassenhagen, I.; Wilken, S.; Godhe, A.; Rengefors, K. Phenotypic plasticity and differentiation in an invasive freshwater microalga. Harmful Algae 2015, 41, 38–45. [Google Scholar] [CrossRef] [Green Version]
- Chung, T.-Y.; Kuo, C.-Y.; Lin, W.-J.; Wang, W.-L.; Chou, J.-Y. Indole-3-acetic-acid-induced phenotypic plasticity in Desmodesmus algae. Sci Rep 2018, 8, 10270. [Google Scholar] [CrossRef]
- Lin, W.-J.; Ho, H.-C.; Chu, S.-C.; Chou, J.-Y. Effects of auxin derivatives on phenotypic plasticity and stress tolerance in five species of the green alga Desmodesmus (Chlorophyceae, Chlorophyta). Peer J. 2020, 8, e8623. [Google Scholar] [CrossRef]
- Giovagnetti, V.; Flori, S.; Tramontano, F.; Lavaud, J.; Brunet, C. The Velocity of Light Intensity Increase Modulates the Photoprotective Response in Coastal Diatoms. PLoS ONE 2014, 9, e103782. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.; Nelson, D.; Yi, Z.; Xu, M.; Khraiwesh, B.; Jijakli, K.; Chaiboonchoe, A.; Alzahmi, A.; Al-Khairy, D.; Brynjolfsson, S.; et al. Bioactive Compounds From Microalgae: Current Development and Prospects. In Studies in Natural Products Chemistry; Rahman, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 199–225. [Google Scholar]
- Sansone, C.; Brunet, C. Promises and Challenges of Microalgal Antioxidant Production. Antioxidants 2019, 8, 199. [Google Scholar] [CrossRef] [Green Version]
- Orefice, I.; Musella, M.; Smerilli, A.; Sansone, C.; Chandrasekaran, R.; Corato, F.; Brunet, C. Role of nutrient concentrations and water movement on diatom’s productivity in culture. Sci. Rep. 2019, 9, 1479. [Google Scholar] [CrossRef] [Green Version]
- Kourtchenko, O.; Rajala, T.; Godhe, A. Growth of a common planktonic diatom quantified using solid medium culturing. Sci. Rep. 2018, 8, 9757. [Google Scholar] [CrossRef]
- Liao, I.C.; Su, H.M.; Lin, J.H. Larval foods for Penaeid prawns. In CRC Handbook of Mariculture, Volume I: Crustacean Aquaculture; McVey, J.P., Ed.; CRC Press: Boca Raton, FL, USA, 1993; pp. 29–59. [Google Scholar]
- Su, H.M.; Lei, C.H.; Liao, I.C. Effect of temperature, illumination and salinity on the growth rates of Skeletonema costatum. J. Fish Soc. Taiwan 1990, 17, 213–222. [Google Scholar]
- Coutteau, P.; Sorgeloos, P. The use of algal substitutes and the requirement for live algae in the hatchery and nursery rearing of bivalve molluscs: An international survey. J. Shellfish Res. 1992, 11, 467–476. [Google Scholar]
- Brown, M.R.; Jeffrey, S.W.; Volkman, J.K.; Dunstan, G.A. Nutritional properties of microalgae for mariculture. Aquaculture 1997, 151, 315–331. [Google Scholar] [CrossRef]
- Miralto, A.; Barone, G.; Romano, G.; Poulet, S.A.; Ianora, A.; Russo, G.L.; Buttino, I.; Mazzarella, G.; Laabir, M.; Cabrini, M.; et al. The insidious effect of diatoms on copepod reproduction. Nature 1999, 402, 173–176. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals; Smith, W.L., Chanley, M.H., Eds.; Springer: Boston, MA, USA, 1975; pp. 29–60. [Google Scholar]
- Brunet, C.; Johnsen, G.; Lavaud, J.; Roy, S. Pigments and photoacclimation processes. In Phytoplankton Pigments: Characterization, Chemotaxonomy and Applications in Oceanography; Roy, S., Llewellyn, C.A., Egeland, E.S., Johnsen, G., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 445–471. [Google Scholar]
- Orefice, I.; Lauritano, C.; Procaccini, G.; Ianora, A.; Romano, G. Insights into possible cell-death markers in the diatom Skeletonema marinoi in response to senescence and silica starvation. Mar. Genomics 2015, 24, 81–88. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Green, L.C.; Wagner, D.A.; Glogowski, J.; Skipper, P.L.; Wishnok, J.S.; Tannenbaum, S.R. Analysis of nitrate, nitrite, and [15N] nitrate in biological fluids. Anal. Biochem. 1982, 126, 131–138. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]




| Light Condition | Ovothiol B Concentration | Glutathione Concentration |
|---|---|---|
| Low sinusoidal light (Sin150) | 50 ± 10 µM | 1.0 ± 0.3 mM |
| High sinusoidal light (Sin600) | 110 ± 20 µM | 2.3 ± 0.3 mM |
| NO | ROS | ovoA | nos1 | nos2 | |
|---|---|---|---|---|---|
| NO | HL, LL | D | - | - | |
| EOS | *** p < 0.001, *** p < 0.001 | - | HL, D | - | |
| ovoA | * p < 0.05 | - | LL | HL, LL | |
| nos1 | - | * p < 0.05, * p < 0.05 | ** p < 0.01 | LL | |
| nos2 | - | - | *** p < 0.001, *** p < 0.001 | ** p < 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milito, A.; Orefice, I.; Smerilli, A.; Castellano, I.; Napolitano, A.; Brunet, C.; Palumbo, A. Insights into the Light Response of Skeletonema marinoi: Involvement of Ovothiol. Mar. Drugs 2020, 18, 477. https://doi.org/10.3390/md18090477
Milito A, Orefice I, Smerilli A, Castellano I, Napolitano A, Brunet C, Palumbo A. Insights into the Light Response of Skeletonema marinoi: Involvement of Ovothiol. Marine Drugs. 2020; 18(9):477. https://doi.org/10.3390/md18090477
Chicago/Turabian StyleMilito, Alfonsina, Ida Orefice, Arianna Smerilli, Immacolata Castellano, Alessandra Napolitano, Christophe Brunet, and Anna Palumbo. 2020. "Insights into the Light Response of Skeletonema marinoi: Involvement of Ovothiol" Marine Drugs 18, no. 9: 477. https://doi.org/10.3390/md18090477
APA StyleMilito, A., Orefice, I., Smerilli, A., Castellano, I., Napolitano, A., Brunet, C., & Palumbo, A. (2020). Insights into the Light Response of Skeletonema marinoi: Involvement of Ovothiol. Marine Drugs, 18(9), 477. https://doi.org/10.3390/md18090477