Computational Assessment of Chito-Oligosaccharides Interactions with Plasma Proteins
Abstract
:1. Introduction
2. Results
2.1. Properties of Small Chito-Oligosaccharides Considered in This Study
2.2. Analysis of the Structural Files of Plasma Proteins
2.3. Molecular Docking Study
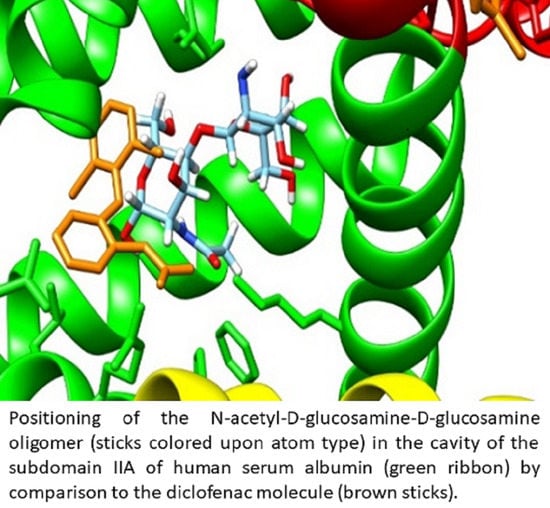
2.4. Characterization of Interactions 0f the Investigated Cos and the Two Plasma Proteins
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Alvarez, F.J. The effect of chitin size, shape, source and purification method on immune recognition. Molecules 2014, 19, 4433–4451. [Google Scholar] [CrossRef] [PubMed]
- Hajji, S.; Younes, I.; Ghorbel-Bellaaj, O.; Hajji, R.; Rinaudo, M.; Nasri, M.; Jellouli, K. Structural differences between chitin and chitosan extracted from three different marine sources. Int. J. Biol. Macromol. 2014, 65, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Raafat, D.; Sahl, H.-G. Chitosan and its antimicrobial potential—A critical literature survey. Microb. Biotechnol. 2009, 2, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.V.R. A review of chitin and chitosan application. React. Funct. Polym. 2000, 46, 1–27. [Google Scholar] [CrossRef]
- Dutta, P.K.; Dutta, J.; Tripath, V.S. Chitin and chitosan: Chemistry, properties and applications. J. Sci. Ind. Res. 2004, 63, 20–31. [Google Scholar]
- Enescu, D.; Olteanu, C.E. Functionalized chitosan and its use in pharmaceutical, biomedical, and biotechnological research. Chem. Eng. Commun. 2008, 195, 1269–1291. [Google Scholar] [CrossRef]
- Hengameh, H.; Mehdi, B. Applications of biopolymers I: Chitosan. Monatshefte Chem. Chem. Mon. 2009, 140, 1403–1420. [Google Scholar]
- Kumar, M.N.V.R.; Muzzarelli, R.A.A.; Muzzarelli, C.; Sashiwa, H.; Domb, A.J. Chitosan chemistry and pharmaceutical perspectives. Chem. Rev. 2004, 104, 6017–6084. [Google Scholar] [CrossRef]
- Elieh-Ali-Komi, D.; Hamblin, M.R. Chitin and chitosan: Production and application of versatile biomedical nanomaterials. Int. J. Adv. Res. 2016, 4, 411–427. [Google Scholar]
- Giri, T.K.; Thakur, A.; Alexander, A.; Ajazuddin; Badwaik, H.; Tripathi, D.K. Modified chitosan hydrogels as drug delivery and tissue engineering systems: Present status and applications. Acta Pharm. Sin. B 2012, 2, 439–449. [Google Scholar] [CrossRef] [Green Version]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H.; Chan, W.Y. Chitosan: An update on potential biomedical and pharmaceutical applications. Mar. Drugs 2015, 13, 5156–5186. [Google Scholar] [CrossRef]
- Patrulea, V.; Hirt-Burri, N.; Jeannerat, A.; Applegate, L.A.; Ostafe, V.; Jordan, O.; Borchard, G. Peptide-decorated chitosan derivatives enhance fibroblast adhesion and proliferation in wound healing. Carbohydr. Polym. 2016, 142, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, K.A.; El-Eswed, B.I.; Abu-Sbeih, K.A.; Arafat, T.A.; Al Omari, M.M.H.; Darras, F.H.; Badwan, A.A. Preparation of chito-oligomers by hydrolysis of chitosan in the presence of zeolite as adsorbent. Mar. Drugs 2016, 14, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaqat, F.; Eltem, R. Chitooligosaccharides and their biological activities: A comprehensive review. Carbohydr. Polym. 2018, 184, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, G.; Kim, Y.S.; Hwang, J.W.; Kim, S.K.; Jeon, Y.J.; Je, J.Y.; Ahn, C.B.; Moon, S.H.; Jeon, B.T.; Park, P.J. Chitooligosac-charide and its derivatives: Preparation and biological applications. Biomed. Res. Int. 2014, 2014, 654913. [Google Scholar] [CrossRef] [Green Version]
- Muanprasat, C.; Chatsudthipong, V. Chitosan oligosaccharide: Biological activities and potential therapeutic applications. Pharmacol. Ther. 2017, 170, 80–97. [Google Scholar] [CrossRef]
- Zou, P.; Yang, X.; Wang, J.; Li, Y.; Yu, H.; Zhang, Y.; Liu, G. Advances in characterisation and biological activities of chitosan and chitosan oligosaccharides. Food Chem. 2016, 190, 1174–1181. [Google Scholar] [CrossRef]
- Roman, D.L.; Roman, M.; Som, C.; Schmutz, M.; Hernandez, E.; Wick, P.; Casalini, T.; Perale, G.; Ostafe, V.; Isvoran, A. Computational assessment of the pharmacological profiles of degradation products of chitosan. Front. Bioeng. Biotechnol. 2019, 7, 214. [Google Scholar] [CrossRef]
- Schönfeld, D.L.; Ravelli, R.B.; Mueller, U.; Skerra, A. The 1.8-Å crystal structure of α1-acid glycoprotein (Orosomucoid) solved by UV RIP reveals the broad drug-binding activity of this human plasma lipocalin. J. Mol. Biol. 2008, 384, 393–405. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, P.; Liang, S.; Zhou, Z.; Wu, X.; Yang, F.; Liang, H. Structural basis of non-steroidal anti-inflammatory drug diclofenac binding to human serum albumin. Chem. Biol. Drug Des. 2015, 86, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. Des. Sel. 1999, 12, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Chadha, N.; Singh, D.; Milton, M.D.; Mishra, G.; Daniel, J.; Mishra, A.K.; Tiwari, A.K. Computational prediction of interaction and pharmacokinetics profile study for polyamino-polycarboxylic ligands on binding with human serum albumin. New J. Chem. 2020, 44, 2907–2918. [Google Scholar] [CrossRef]
- Lambrinidis, G.; Vallianatou, T.; Tsantili-Kakoulidou, A. In Vitro, in silico and integrated strategies for the estimation of plasma protein binding. A review. Adv. Drug Deliv. Rev. 2015, 86, 27–45. [Google Scholar] [CrossRef]
- Vuignier, K.; Schappler, J.; Veuthey, J.-L.; Carrupt, P.-A.; Martel, S. Drug–protein binding: A critical review of analytical tools. Anal. Bioanal. Chem. 2010, 398, 53–66. [Google Scholar] [CrossRef]
- Chemicalize.com Home Page. Available online: http://chemicalize.com (accessed on 7 June 2020).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visu-alization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 3, W270–W277. [Google Scholar] [CrossRef] [Green Version]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef]
- Protein-Ligand Interaction Profiler Home Page. Available online: https://projects.biotec.tu-dresden.de/plip-web/plip/index (accessed on 20 January 2021).
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein–Ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]







| Deacetilation Degree | Deacetylation Pattern | Acronym | MW (g/mol) |
|---|---|---|---|
| 0% | GlcNAc | 1A | 221.21 |
| (GlcNAc)2 | 2A | 424.40 | |
| (GlcNAc)3 | 3A | 627.59 | |
| (GlcNAc)4 | 4A | 870.79 | |
| (GlcNAc)5 | 5A | 1033.98 | |
| (GlcNAc)6 | 6A | 1237.17 | |
| 33% | GlcNAc-GlcN-GlcNAc | ADA | 585.56 |
| 50% | GlcN-GlcNAc | DA | 382.36 |
| (GlcN-GlcNAc)2 | DADA | 746.71 | |
| (GlcNAc-GlcN)2 | ADAD | 746.71 | |
| GlcNAc-GlcNAc-GlcN-GlcN | AADD | 746.71 | |
| GlcN-GlcN-GlcNAc-GlcNAc | DDAA | 746.71 | |
| GlcN-GlcNAc-GlcNAc-GlcN | DAAD | 746.71 | |
| GlcNAc-GlcN-GlcN-GlcNAc | ADDA | 746.71 | |
| (GlcN-GlcNAc)3 | DADADA | 1475.41 | |
| (GlcNAc-GlcN)3 | ADADAD | 1475.41 | |
| 67% | GlcN-GlcN-GlcNAc | DDA | 543.52 |
| GlcNAc-GlcN-GlcN-GlcN-GlcNAc-GlcN | ADDDAD | 1069.02 | |
| GlcN-GlcN-GlcN-GlcNAc-GlcN-GlcNAc | DDDADA | 1069.02 | |
| 100% | GlcN | D | 179.17 |
| (GlcN)2 | 2D | 340.33 | |
| (GlcN)3 | 3D | 501.48 | |
| (GlcN)4 | 4D | 662.64 | |
| (GlcN)5 | 5D | 823.79 | |
| (GlcN)6 | 6D | 984.95 |
| COs | Contacts with Alpha-1-Glycoprotein (AGP) | Contacts with Human Serum Albumin (HSA) | ||||||
|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) | Hydrophobic Interactions | Hydrogen Bonds | Salt Bridges | Binding Energy (kcal/mol) | Hydrophobic Interactions | Hydrogen Bonds | Salt Bridges | |
| 1A | −5.90 | PHE49 | SER89, ARG90, HIS97 | ARG90 | −6.70 | LEU219 | ARG257, SER287 | none |
| 1D | −5.47 | none | TYR27, TYR127 | ARG90 | −4.89 | none | ARG257 (2) | none |
| 2A | −8.11 | PHE49 | SER30, GLU64, GLN66, ARG90, SER125 (2) | none | −8.69 | LEU238, VAL241 | LYS199, ARG218, ARG257, ILE290 | none |
| 2D | −7.66 | none | ARG90 | HIS97 | −8.37 | none | LYS199, ARG257 (3), SER287 | LYS199, HIS242 |
| DA | −7.94 | PHE114 | GLN66, ARG90 (2), SER125 (2), TYR127 | none | −8.42 | LEU219, ARG222, PHE223 | LYS199, HIS242, ARG257, SER287 | ARG257 |
| 3A | −8.75 | TYR27, TYR37, ARG90, HIS97, PHE114 (2) | TYR37 (2), GLN66, ARG90, HIS97 (2) | ARG90, HIS97 | −8.44 | ARG218, LEU219, LEU238 | SER192, LYS195, GLN196, LYS199, ARG218 (2), ARG257 | LYS199 |
| ADA | −8.15 | ILE88, LEU112 | GLU64, GLN66, ARG90 (2), HIS97, TYR110 (2) | ARG90 | −10.28 | GLN196, ALA261 | GLU153, ARG218 (2), ARG257 (2) | LYS199 (2) |
| DDA | −8.74 | ILE88, ALA99 | SER40, ARG90, TYR127 | ARG90 | −9.89 | none | ARG257, SER287, ALA291, GLU292 (2) | LYS195, LYS199, ARG257 |
| 3D | −5.36 | none | GLU36, SER40 | none | −5.74 | none | TYR148, TYR150, GLN196 (2), GLU292 | LYS195, ARG257 |
| 4A | −10.44 | VAL41, ILE44, ILE88, GLN95, LEU112 | GLU36, SER40, ARG90, GLY93, HIS97 (4) | ARG90, HIS97 | −10.53 | LEU260 | GLU153, SER192, LYS195, GLN196, LYS199, ARG218, ARG257 (2), HIS288 | LYS199, HIS288 |
| AADD | −9.36 | TYR27, ARG90, PHE114 | SER40, ARG90, HIS97 (2), SER125 (2) | ARG90 | −10.51 | none | LYS195, ARG218, ARG222 (2), ARG257, SER287, ASP451 | LYS195, LYS199, ARG218 |
| ADAD | −9.56 | TYR37, ARG90, PHE114 | GLU36 (2), SER40 (2), THR47, GLN66, ARG90 | none | −10.18 | TYR452 | GLU153, ARG160, GLU188, ARG218 (2), ARG222, ASP451 | ARG160, LYS195, LYS281, HIS288 |
| ADDA | −9.82 | TYR27 | SER40 (2), GLU64 (2), ARG90, HIS97 | none | −11.62 | GLU153, PHE157 | ARG160, SER192, GLN196 (2), GLU292 (3) | LYS195, LYS199 (2), ARG257 |
| DAAD | −8.31 | ARG90, PHE114 | SER30 (2), GLU36 (3), SER40, THR47, GLN66, ARG90, SER125 (2), TYR127 | none | −7.32 | GLN196 | GLU153, LYS199, ARG218, HIS242, ARG257 (2), HIS288 (2) | LYS195, LYS199, ARG222 |
| DADA | −6.11 | ARG90, VAL92, PHE114 | GLU64, ARG68 (2), ARG90, SER125, TYR127 | ARG90 | −6.89 | PHE156 | ARG160 (2), GLU184, GLU188, HIS288 (2), GLU292 | ARG160 (2) |
| DDAA | −9.32 | PHE49, PHE51, LEU112, TYR127 | SER40 (3), ARG68 (2), HIS97, TYR127 | ARG90 | −10.34 | TYR148, GLN196, ARG197 | GLN196, LYS199, ARG257 | ARG257 |
| 4D | −9.85 | none | GLU36, SER40 (2), GLU64, ARG90, SER125 (2), TYR127 | none | −11.25 | none | GLU6, ARG10, GLU252 (3), ASP255 | HIS3, LYS240 |
| 5A | −10.54 | TYR37, ILE88, ALA99, LEU112 | GLU36 (2), SER40, GLU64, ARG90, GLY93, HIS97 | none | −13.53 | LEU260 | ARG160, LYS195, LYS199, ARG218 (2), ARG222, ARG257, ALA291, GLU292 (2), TYR452 | LYS195, LYS199 |
| 5D | −6.77 | none | TYR27, SER30, GLU36, SER40, ARG90, HIS97, SER125, TYR127 | none | −6.81 | none | TYR148, GLU153 (3), GLU188, SER192 (2), LYS199 (2), HIS242 (2), GLU292 (2) | ARG160, LYS195, LYS199 |
| 6A | −11.08 | TYR37, ILE44, LEU79, ILE88, ARG90, PHE114 | TYR37, SER40 (2), GLU64, ARG68 (2), ARG90, HIS97 (2), TYR127 | ARG68, ARG90 (2) | −12.07 | ALA191, GLN196 | GLU153 (2), LYS195, LYS199, ARG218 (2), ARG222 (2), ARG257, HIS288 (3), GLU292 (3) | ARG160, LYS195, LYS199, LYS281, HIS288 |
| ADADAD | −10.31 | PHE32, TYR37, ARG90, VAL92, PHE114 | SER30, GLU36 (2), TYR37, SER40 (4), GLU64, GLN66, ARG90, GLY93, HIS97 (2), SER125 (2) | none | −13.03 | THR420 | GLU505, THR506, HIS510, LYS524 (2), THR527 | LYS524 (3) |
| DADADA | −9.62 | TYR27, TYR37, ILE44 | SER40, GLN66, ARG68 (2), TYR127 | ARG90 (2) | −9.68 | LEU260, ALA261 | ARG160, SER192, LYS195, LYS199 (2), ARG218 (2), ARG222, ARG257 (2), GLU292 (3), VAL293 | LYS195, LYS199 |
| ADDDAD | −8.27 | ALA99 | GLU36, GLU43 | ARG90 | −8.34 | LYS436, TYR452 | GLU184, GLU188, HIS288 (2), GLU292 (3), LYS436 (2), TYR452 (2) | ARG160, LYS436 |
| DDDADA | −8.21 | PHE32, ALA99, PHE114 | GLU36, SER40, GLN66, ARG90, SER125 (2), TYR127 | none | −8.28 | LEU260 | GLU153, LYS199 (2), ARG218, SER287, HIS288 (3), ALA291, GLU292 (3) | LYS195, ARG257 |
| 6D | −9.41 | none | GLU36, SER40 (2), GLU64, ARG90, ASN117, ASP118, ASN121 | ARG90, HIS97 | −13.74 | none | GLU153 (2), ARG160 (2), GLU188, LYS195 (2), LYS199, ARG218, GLU292 | ARG160, LYS195, LYS199 (2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roman, D.L.; Ostafe, V.; Isvoran, A. Computational Assessment of Chito-Oligosaccharides Interactions with Plasma Proteins. Mar. Drugs 2021, 19, 120. https://doi.org/10.3390/md19030120
Roman DL, Ostafe V, Isvoran A. Computational Assessment of Chito-Oligosaccharides Interactions with Plasma Proteins. Marine Drugs. 2021; 19(3):120. https://doi.org/10.3390/md19030120
Chicago/Turabian StyleRoman, Diana Larisa, Vasile Ostafe, and Adriana Isvoran. 2021. "Computational Assessment of Chito-Oligosaccharides Interactions with Plasma Proteins" Marine Drugs 19, no. 3: 120. https://doi.org/10.3390/md19030120
APA StyleRoman, D. L., Ostafe, V., & Isvoran, A. (2021). Computational Assessment of Chito-Oligosaccharides Interactions with Plasma Proteins. Marine Drugs, 19(3), 120. https://doi.org/10.3390/md19030120