Antarctic Thraustochytrids as Sources of Carotenoids and High-Value Fatty Acids
Abstract
:1. Introduction
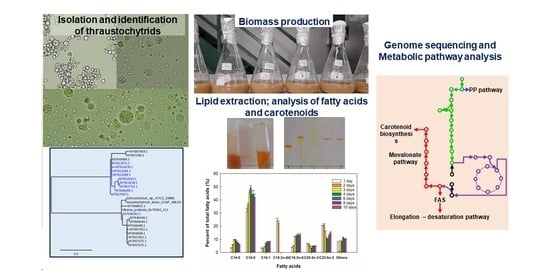
2. Results
2.1. Lipid and Carotenoid Accumulation
2.2. Thraustochytrium sp. RT2316-16
2.3. Thraustochytrium sp. RT2316-16 Genome Sequencing
2.4. Biosynthesis of Fatty Acids in Thraustochytrium sp. RT2316-16
2.5. Carotenoid Biosynthesis by Thraustochytrium sp. RT2316-16
3. Discussion
4. Materials and Methods
4.1. Collection and Isolation of Thraustochytrids
4.2. DNA Extraction and Molecular Identification of Isolates
4.3. Genome Sequencing and Assembly
4.4. Data Availability
4.5. Gene Prediction and Annotation
4.6. Production of Biomass, Lipids and Carotenoids
4.7. Concentrations of Biomass, Residual Sugars and Amino Acids
4.8. Extraction of Total Lipids and Fatty Acid Profile Determination
4.9. Extraction and Quantification of Carotenoids
4.10. Thin Layer Chromatography (TLC)
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Bazan, N.G.; Molina, M.F.; Gordon, W.C. Docosahexaenoic acid signalolipidomics in nutrition: Significance in aging, neuroinflammation, macular degeneration, Alzheimer’s, and other neurodegenerative diseases. Annu. Rev. Nutr. 2011, 31, 321–351. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef]
- Mason, R.P.; Libby, P.; Bhatt, D.L. Emerging mechanisms of cardiovascular protection for the omega-3 fatty acid eicosapentaenoic acid. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1135–1147. [Google Scholar] [CrossRef]
- Kalkman, H.O.; Hersberger, M.; Walitza, S.; Berger, G.E. Disentangling the molecular mechanisms of the antidepressant activity of omega-3 polyunsaturated fatty acid: A comprehensive review of the literature. Int. J. Mol. Sci. 2021, 22, 4393. [Google Scholar] [CrossRef]
- Stahl, W.; Sies, H. Bioactivity and protective effects of natural carotenoids. Biochim. Biophys. Acta 2005, 1740, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pino-Maureira, N.L.; González-Saldía, R.R.; Capdeville, A.; Srain, B. Rhodotorula strains isolated from seawater that can biotransform raw glycerol into docosahexaenoic acid (DHA) and carotenoids for animal nutrition. Appl. Sci. 2021, 11, 2824. [Google Scholar] [CrossRef]
- Romari, K.; Godart, F.; Calleja, P. Production of Docosahexaenoic Acid and/or Eicosapentaenoic Acid and/or Carotenoids in Mixotrophic Mode by Nitzschia. U.S. Patent 0044737 A1, 21 February 2015. [Google Scholar]
- Chang, M.; Zhang, T.; Li, L.; Lou, F.; Ma, M.; Liu, R.; Jin, Q.; Wang, X. Choreography of multiple omics reveals the mechanism of lipid turnover in Schizochytrium sp. S31. Algal Res. 2021, 54, 102182. [Google Scholar] [CrossRef]
- Ward, O.P.; Singh, A. Omega-3/6 fatty acids: Alternative sources of production. Process Biochem. 2005, 40, 3627–3652. [Google Scholar] [CrossRef]
- Chang, K.J.L.; Mansour, M.P.; Dunstan, G.A.; Blackburn, S.I.; Koutoulis, A.; Nichols, P.D. Odd-chain polyunsaturated fatty acids in thraustochytrids. Phytochemistry 2011, 72, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Sakaguchi, K.; Matsuda, T.; Abe, E.; Hama, Y.; Hayashi, M.; Honda, D.; Okita, Y.; Sugimoto, S.; Okino, N.; et al. Increase of eicosapentaenoic acid in thraustochytrids through thraustochytrid ubiquitin promoter-driven expression of a fatty acid Δ5 desaturase gene. Appl. Environ. Microbiol. 2011, 77, 3870–3876. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wang, T. Characterization of lipid components in two microalgae for biofuel application. J. Am. Oil Chem. Soc. 2012, 89, 135–143. [Google Scholar] [CrossRef]
- Aki, T.; Hachida, K.; Yoshinaga, M.; Katai, Y.; Yamasaki, T.; Kawamoto, S.; Kakizono, T.; Maoka, T.; Shigeta, S.; Suzuki, O.; et al. Thraustochytrid as a potential source of carotenoids. J. Am. Oil. Chem. Soc. 2003, 80, 789–794. [Google Scholar] [CrossRef]
- Carmona, M.L.; Naganuma, T.; Yamaoka, Y. Identification by HPLC-MS of carotenoids of the Thraustochytrium CHN-1 strain isolated from the Seto Inland Sea. Biosci. Biotechnol. Biochem. 2003, 67, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, T.; Aki, T.; Shinozaki, M.; Taguchi, M.; Kawamoto, S.; Ono, K. Utilization of Shochu distillery wastewater for production of polyunsaturated fatty acids and xanthophylls using thraustochytrid. J. Biosci. Bioeng. 2006, 102, 323–327. [Google Scholar] [CrossRef]
- Quilodrán, B.; Hinzpeter, I.; Hormazabal, E.; Quiroz, A.; Shene, C. Docosahexaenoic acid (C22:6n−3, DHA) and astaxanthin production by Thraustochytriidae sp. AS4-A1 a native strain with high similitude to Ulkenia sp.: Evaluation of liquid residues from food industry as nutrient sources. Enzyme Microb. Technol. 2010, 14, 24–30. [Google Scholar] [CrossRef]
- Park, H.; Kwak, M.; Seo, J.; Ju, J.H.; Heo, S.Y.; Park, S.M.; Hong, W.K. Enhanced production of carotenoids using a Thraustochytrid microalgal strain containing high levels of docosahexaenoic acid-rich oil. Bioprocess Biosyst. Eng. 2018, 41, 1355–1370. [Google Scholar] [CrossRef]
- Bowles, R.D.; Hunt, A.E.; Bremer, G.B.; Duchars, M.G.; Eaton, R.A. Long-chain n−3 polyunsaturated fatty acid production by members of the marine protistan group the thraustochytrids: Screening of isolates and optimisation of docosahexaenoic acid production. J. Biotechnol. 1999, 70, 193–202. [Google Scholar] [CrossRef]
- Paredes, P.; Larama, G.; Flores, L.; Leyton, A.; Ili, C.G.; Asenjo, J.A.; Chisti, Y.; Shene, C. Temperature differentially affects gene expression in Antarctic thraustochytrid Oblongichytrium sp. RT2316-13. Mar. Drugs 2020, 18, 563. [Google Scholar] [CrossRef] [PubMed]
- Shene, C.; Paredes, P.; Flores, L.; Leyton, A.; Asenjo, J.A.; Chisti, Y. Dynamic flux balance analysis of biomass and lipid production by Antarctic thraustochytrid Oblongichytrium sp. RT2316-13. Biotechnol. Bioeng. 2020, 117, 3006–3017. [Google Scholar] [CrossRef]
- Yokoyama, R.; Honda, D. Taxonomic rearrangement of the genus Schizochytrium sensu lato based on morphology, chemotaxonomic characteristics, and 18S rRNA gene phylogeny (Thraustochytriaceae, Labyrinthulomycetes): Emendation for Schizochytrium and erection of Aurantiochytrium and Oblongichytrium gen. nov. Mycoscience 2007, 48, 199–211. [Google Scholar] [CrossRef]
- Dieser, M.; Greenwood, M.; Foreman, C.M. Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef] [Green Version]
- Dellero, Y.; Cagnac, O.; Rose, S.; Seddiki, K.; Cussac, M.; Morabito, C.; Lupette, J.; Cigliano, R.A.; Sanseverino, W.; Kuntz, M.; et al. Proposal of a new thraustochytrid genus Hondaea gen. nov. and comparison of its lipid dynamics with the closely related pseudo-cryptic genus Aurantiochytrium. Algal Res. 2018, 35, 125–141. [Google Scholar] [CrossRef]
- Iwasaka, H.; Koyanagi, R.; Satoh, R.; Nagano, A.; Watanabe, K.; Hisata, K.; Satoh, N.; Aki, T. A possible trifunctional-carotene synthase gene identified in the draft genome of Aurantiochytrium sp. strain KH105. Genes 2018, 9, 200. [Google Scholar] [CrossRef] [Green Version]
- Marchan, L.F.; Chang, K.J.L.; Nichols, P.D.; Mitchell, W.J.; Polglase, J.L.; Gutierrez, T. Taxonomy, ecology and biotechnological applications of thraustochytrids: A review. Biotechnol. Adv. 2018, 36, 26–46. [Google Scholar] [CrossRef]
- Liu, B.; Ertesvåg, H.; Aasen, J.M.; Vadstein, O.; Brautaset, T.; Heggeset, T.M.B. Draft genome sequence of the docosahexaenoic acid producing Thraustochytrid Aurantiochytrium sp. T66. Genom. Data 2016, 8, 115–116. [Google Scholar] [CrossRef] [Green Version]
- Sediki, K.; Godart, F.; Cigliano, R.; Sanseverino, W.; Barakata, M.; Ortet, P.; Rébellié, F.; Maréchal, E.; Cagnac, O.; Amato, A. Sequencing, de novo assembly, and annotation of the complete genome of a new Thraustochytrid species, strain CCAP_4062/3. Genome Annunc. 2018, 6, e01335-17. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Liu, M.; He, M.; Ye, Y.; Huang, J. Illustrating and enhancing the biosynthesis of astaxanthin and docosahexaenoic acid in Aurantiochytrium sp. SK4. Mar. Drugs 2019, 17, 45. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.J.; Mo, K.Q.; Ren, L.J.; Li, G.L.; Huang, J.Z.; Haung, H. Genome sequence of Schizochytrium sp. CCTCC M209059, an effective producer of docosahexaenoic acid-rich lipids. Genome Announc. 2015, 3, e00819-15. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Dauenpen, M.; Qu, X.; Qi, X. Genomic analysis of genes involved in the biosynthesis of very long chain polyunsaturated fatty acids in Thraustochytrium sp. 26185. Lipids 2016, 51, 1065–1075. [Google Scholar] [CrossRef]
- Song, Z.; Stajich, J.E.; Xie, Y.; Liu, X.; He, Y.; Chen, J.; Hicks, G.R.; Wang, G. Comparative analysis reveals unexpected genome features of newly isolated Thraustochytrids strains: On ecological function and PUFAs biosynthesis. BMC Genom. 2018, 17, 541. [Google Scholar] [CrossRef]
- Metz, J.G.; Roessler, P.; Facciotti, D.; Levering, C.; Dittrich, F.; Lassner, M.; Valentine, R.; Lardizabal, K.; Domergue, F.; Yamada, A.; et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 2001, 293, 290–293. [Google Scholar] [CrossRef]
- Zhu, X.; Li, S.; Liu, L.; Li, S.; Luo, Y.; Lv, C.; Wang, B.; Cheng, C.H.K.; Chen, H.; Yang, X. Genome sequencing and analysis of Thraustochytriidae sp. SZU445 provides novel insights into the polyunsaturated fatty acid biosynthesis pathway. Mar. Drugs 2020, 18, 118. [Google Scholar] [CrossRef] [Green Version]
- Kihara, A. Very long-chain fatty acids: Elongation, physiology and related disorders. J. Biochem. 2012, 152, 387–395. [Google Scholar] [CrossRef]
- Moon, Y.A.; Horton, J.D. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J. Biol. Chem. 2003, 278, 7335–7343. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.J.; Huang, J.C.; Liu, J.; Li, Y.; Jiang, Y.; Xu, Z.F.; Sandmann, G.; Chen, F. Functional characterization of various algal carotenoid ketolases reveals that ketolating zeaxanthin efficiently is essential for high production of astaxanthin in transgenic Arabidopsis. J. Exp. Bot. 2011, 62, 3659–3669. [Google Scholar] [CrossRef]
- Ojima, K.; Breitenbach, J.; Visser, H.; Setoguchi, Y.; Tabata, K.; Hoshino, T.; van den Berg, J.; Sandmann, G. Cloning of the astaxanthin synthase gene from Xanthophyllomyces dendrorhous (Phaffia rhodozyma) and its assignment as a β-carotene 3-hydroxylase/4-ketolase. Mol. Gen. Genomics 2006, 275, 148–158. [Google Scholar] [CrossRef]
- Shene, C.; Leyton, A.; Rubilar, M.; Pinelo, M.; Acevedo, F.; Morales, E. Production of lipids and docosahexasaenoic acid (DHA) by a native Thraustochytrium strain. Eur. J. Lipid Sci. Technol. 2013, 115, 890–900. [Google Scholar] [CrossRef]
- Mo, C.; Douek, J.; Rinkevich, B. Development of a PCR strategy for thraustochytrid identification based on 18S rDNA sequence. Mar. Biol. 2002, 140, 883–889. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Krueger, F. Trim Galore! A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. 2015. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 1 March 2020).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 March 2020).
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L.; et al. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 27, 768–777. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Holt, C.; Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011, 12, 491. [Google Scholar] [CrossRef] [Green Version]
- Slater, G.S.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef] [Green Version]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open-source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef] [PubMed]
- Hass, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef] [Green Version]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrêa Dos Santos, A.R.; Goldman, G.H.; Riaño-Pachón, D.M. ploidyNGS: Visually exploring ploidy with Next Generation Sequencing data. Bioinformatics 2017, 33, 2575–2576. [Google Scholar] [CrossRef]
- Bertrand-Harb, C.; Nicolas, M.G.; Dalgalarraondo, M.; Chobert, J.M. Determination of alkylation degree by three colorimetric methods and amino-acid analysis: A comparative study. Sci. Aliment. 1993, 13, 577–584. [Google Scholar]
- Bligh, E.; Dyer, W. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Vijayalakshmi, G.; Shobha, B.; Vanajakshi, V.; Divakar, S.; Manohar, B. Response surface methodology for optimization of growth parameters for the production of carotenoids by a mutant strain of Rhodotorula gracilis. Eur. Food Res. Technol. 2001, 213, 234–239. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]


| Strain | Closest Relative in GenBank | Identity Match (%) | Accession | X (g/L) | TL (%) | EPA (%) | DHA (%) |
|---|---|---|---|---|---|---|---|
| RT2316-14 | Oblongichytrium sp. | 98.45 | MT648446 | 2.7 ± 0.0 b | 16.0 ± 1.3 a,b | 9.9 ± 0.3 d | 26.1 ± 1.2 d |
| RT2316-15 | Oblongichytrium sp. | 99.32 | MT648448 | 3.7 ± 0.2 a | 16.1 ± 0.5 a,b | 8.4 ± 0.2 e | 21.6 ± 1.7 e |
| RT2316-18 | Thraustochytrium sp. | 100.00 | MT648499 | 2.7 ± 0.1 b | 12.3 ± 0.3 b,c | 7.1 ± 0.5 f | 20.8 ± 1.2 e |
| RT2316-21 | Oblongichytrium sp. | 99.47 | MT667280 | 2.0 ± 0.2 c | 9.9 ± 1.0 c | 10.4 ± 1.9 d | 22.0 ± 1.0 e |
| RT2316-22 | Oblongichytrium sp. | 98.92 | MT667372 | 1.3 ± 0.2 d | 9.3 ± 0.1 c | 9.1 ± 0.0 d | 22.8 ± 0.0 e |
| RT2316-23 | Oblongichytrium sp. | 99.46 | MT667375 | 1.8 ± 0.1 c | 9.8 ± 1.2 c | 13.3 ± 0.1 c | 37.4 ± 1.9 a |
| RT2316-24 | Oblongichytrium sp. | 99.46 | MT667352 | 3.8 ± 0.1 a | 13.4 ± 0.2 b | 1.3 ± 0.0 g | 2.1 ± 0.0 f |
| RT2316-25 | Oblongichytrium sp. | 99.19 | MT667384 | 2.2 ± 0.2 c | 14.1 ± 1.0 b | 13.2 ± 0.2 c | 29.5 ± 0.9 c |
| RT2316-26 | Oblongichytrium sp. | 98.81 | MT667421 | 3.9 ± 0.1 a | 10.2 ± 0.1 c | 18.2 ± 1.0 a | 39.2 ± 2.8 a |
| RT2316-28 | Aurantiochytrium sp. | 98.34 | MT668501 | 1.2 ± 0.1 d | 15.9 ± 1.4 a,b | ND | ND |
| RT2316-29 | Oblongichytrium sp. | 99.78 | MT668508 | 2.9 ± 0.0 b | 13.8 ± 0.9 b | 8.2 ± 0.0 e | 28.8 ± 0.0 c |
| RT2316-31 | Oblongichytrium sp. | 98.24 | MT668540 | 3.0 ± 0.2 b | 17.3 ± 0.8 a | 14.8 ± 0.3 b | 33.8 ± 2.2 b |
| Strain | Closest Relative in GenBank | Identity Match (%) | Accession | X (g/L) | TL (%) | TC (µg/g) | EPA (%) | DHA (%) |
|---|---|---|---|---|---|---|---|---|
| RT2316-37 | Thraustochytrium sp. | 99.48 | MT812689 | 7.1 ± 0.3 b | 21.6 ± 0.8 b,c | 39.9 ± 1.3 c,d | 1.5 ± 0.0 b | 3.3 ± 0.4 b,c |
| RT2316-38 | Thraustochytrium sp. | 99.25 | MT812701 | 1.9 ± 0.1 d | 23.2 ± 1.2 b | 27.4 ± 5.3 e | 6.7 ± 1.1 a | 50.2 ± 2.7 a |
| RT2316-16 | Thraustochytrium sp. | 100.00 | MT648462 | 7.2 ± 0.2 b | 26.3 ± 1.7 a | 63.9 ± 3.2 a | 6.0 ± 0.0 a | 4.2 ± 0.0 b |
| RT2316-45 | Thraustochytrium sp. | 99.45 | MT814239 | 7.3 ± 0.1 b | 18.6 ± 1.2 c | 35.2 ± 6.6 d | 2.6 ± 1.5 b | 5.2 ± 1.5 b |
| RT2316-44 | Thraustochytrium sp. | 99.26 | MT814238 | 8.3 ± 0.2 a | 20.1 ± 1.3 b,c | 30.3 ± 2.7 d | 1.9 ± 0.0 b | 4.3 ± 0.4 b |
| RT2316-17 | Thraustochytrium sp. | 95.33 | MT648465 | 5.8 ± 0.3 c | 18.0 ± 0.3 c | 46.1 ± 3.2 c | 0.9 ± 0.5 b | 2.4 ± 1.2 c |
| RT2316-42 | Thraustochytrium sp. | 99.37 | MT812971 | 1.1 ± 0.1 f | 14.3 ± 2.1 d | 54.6 ± 1.7 b | 1.5 ± 0.5 b | 4.1 ± 0.0 b |
| RT2316-40 | Thraustochytrium sp. | 99.34 | MT812947 | 1.5 ± 0.1 e | 18.1 ± 0.5 c | 43.7 ± 1.3 c | 7.1 ± 0.0 a | 47.5 ± 0.0 a |
| RT2316-49 | Thraustochytriidae sp. | 99.47 | MT814293 | 8.3 ± 0.1 a | 18.5 ± 0.7 c | 40.5 ± 2.0 c,d | 1.6 ± 0.2 b | 4.1 ± 0.4 b |
| RT2316-50 | Aurantiochytrium sp. | 99.27 | MT814296 | 5.8 ± 0.3 c | 19.0 ± 0.2 c,d,e | ND | 0.8 ± 0.0 b | 2.7 ± 0.4 c |
| Parameter | Value |
|---|---|
| Number of contigs | 9532 |
| Contigs ≥ 1000 bp | 5133 |
| Contigs ≥ 5000 bp | 2074 |
| Contigs ≥ 10,000 bp | 1167 |
| Contigs ≥ 25,000 bp | 378 |
| Largest contig | 344,669 bp |
| Total length | 44,309,409 bp |
| N50 | 17,465 |
| L50 | 601 |
| Guanine-cytosine content (%) | 58.85 |
| N’s per 100 kbp | 439.38 |
| Parameter | Value |
|---|---|
| Complete BUSCOs | 194 (76.1%) |
| Single-copy BUSCOs | 191 (74.9%) |
| Duplicated BUSCOs | 3 (1.2%) |
| Fragmented BUSCOs | 22 (8.6%) |
| Missing BUSCOs | 39 (15.3%) |
| Total BUSCO genes | 255 (100%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leyton, A.; Flores, L.; Shene, C.; Chisti, Y.; Larama, G.; Asenjo, J.A.; Armenta, R.E. Antarctic Thraustochytrids as Sources of Carotenoids and High-Value Fatty Acids. Mar. Drugs 2021, 19, 386. https://doi.org/10.3390/md19070386
Leyton A, Flores L, Shene C, Chisti Y, Larama G, Asenjo JA, Armenta RE. Antarctic Thraustochytrids as Sources of Carotenoids and High-Value Fatty Acids. Marine Drugs. 2021; 19(7):386. https://doi.org/10.3390/md19070386
Chicago/Turabian StyleLeyton, Allison, Liset Flores, Carolina Shene, Yusuf Chisti, Giovanni Larama, Juan A. Asenjo, and Roberto E. Armenta. 2021. "Antarctic Thraustochytrids as Sources of Carotenoids and High-Value Fatty Acids" Marine Drugs 19, no. 7: 386. https://doi.org/10.3390/md19070386
APA StyleLeyton, A., Flores, L., Shene, C., Chisti, Y., Larama, G., Asenjo, J. A., & Armenta, R. E. (2021). Antarctic Thraustochytrids as Sources of Carotenoids and High-Value Fatty Acids. Marine Drugs, 19(7), 386. https://doi.org/10.3390/md19070386