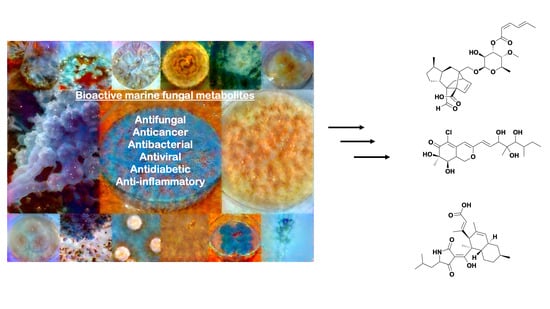
Extremophilic Fungi from Marine Environments: Underexplored Sources of Antitumor, Anti-Infective and Other Biologically Active Agents
Abstract
:1. Introduction
1.1. Antitumor Agents from Deep-Sea Sediments
1.2. Indian Ocean
1.3. Seas near China
1.4. Eastern Pacific Ocean
2. Anti-Infective Agents from Deep-Sea Fungi
2.1. South Atlantic Ocean
2.2. South China Sea
2.3. Indian Ocean
2.4. West Pacific Ocean
3. Other Biologically Active Agents from Deep-Sea Fungi
3.1. (Probably) South China Sea
3.2. Antarctica
4. Bioactive Compounds from Fungal Endophytes of Mangroves
5. Preclinical and Clinical Trials of Mangrove Endophytic-Sourced Compounds
6. Plinabulin Clinical Trials
7. Bioactive Compounds from Fungi Isolated from Marine Vertebrates and Selected Invertebrates
8. Further Piscine-Sourced Agents
9. Bioactive Compounds from Fungi from Marine Invertebrates, Seagrass, Cyanobacteria and Algae
10. Bioactive Compounds from Epigenetic Modifications
11. OSMAC
12. Optimization of Growth Conditions and Strains for Large-Scale Production
13. Materials Produced by Both Terrestrial and Marine Isolates
14. Conclusions
Funding
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giddings, L.-A.; Newman, D.J. Bioactive compounds from marine extremophiles. In Extremophilic Bacteria, Springer Briefs in Microbiology; Tiquia-Arashiro, S.M., Mormile, M., Eds.; Springer: Cham, Switzerland, 2015; pp. 1–150. [Google Scholar] [CrossRef]
- Schulz, B.; Draeger, S.; de la Cruz, T.E.; Rheinheimer, J.; Siems, K.; Loesgen, S.; Bitzer, J.; Schloerke, O.; Zeeck, A.; Kock, I.; et al. Screening strategies for obtaining novel, biologically active, fungal secondary metabolites from marine habitats. Bot. Mar. 2008, 51, 219–234. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism-from biochemistry to genomics. Nat. Rev. Micro. 2005, 3, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.P.; Rama, T.; Oosterhuis, R.; Walker, A.K.; Panf, K.-L. The neglected marine fungi, sensu stricto, and their isolation for natural products’ discovery. Mar. Drugs 2019, 17, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giddings, L.-A.; Newman, D.J. Bioactive compounds from extremophilic marine fungi. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 349–382. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Y.; Li, S.; Wang, Q.; Hu, C.; Liu, H.; Zhang, W. Bioactive metabolites from the deep-sea-derived fungus Diaporthe longicolla fs429. Mar. Drugs 2020, 18, 381. [Google Scholar] [CrossRef]
- Zhang, Z.; He, X.; Liu, C.; Che, Q.; Zhu, T.; Gu, Q.; Li, D. Clindanones A and B and cladosporols F and G, polyketides from the deep-sea derived fungus Cladosporium cladosporioides HDN14-342. RSC Adv. 2016, 6, 76498–76504. [Google Scholar] [CrossRef]
- Li, H.-L.; Li, X.-M.; Mándi, A.; Antus, S.; Li, X.; Zhang, P.; Liu, Y.; Kurtán, T.; Wang, B.-G. Characterization of cladosporols from the marine algal-derived endophytic fungus Cladosporium cladosporioides EN-399 and configurational revision of the previously reported cladosporol derivatives. J. Org. Chem. 2017, 82, 9946–9954. [Google Scholar] [CrossRef]
- Xu, J.-L.; Liu, H.-X.; Chen, Y.-C.; Tan, H.-B.; Guo, H.; Xu, L.-Q.; Li, S.-N.; Huang, Z.-L.; Li, H.-H.; Gao, X.-X.; et al. Highly substituted benzophenone aldehydes and eremophilane derivatives from the deep-sea derived fungus Phomopsis lithocarpus FS508. Mar. Drugs 2018, 16, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Guo, W.; He, X.; Che, Q.; Zhu, T.; Gu, Q.; Li, D. Peniphenylanes A–G from the deep-sea-derived fungus Penicillium fellutanum HDN14-323. Planta Med. 2016, 82, 872–876. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.-W.; Liu, H.-X.; Sun, Z.-H.; Chen, Y.-C.; Tan, Y.-Z.; Zhang, W.-M. Secondary metabolites from the deep-sea derived fungus Acaromyces ingoldii FS121. Molecules 2016, 21, 371. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Li, S.; Chen, Z.; Li, Z.; Liao, Y.; Chen, J. Secondary metabolites produced by the deep-sea-derived fungus Engyodontium album. Chem. Nat. Compds. 2017, 53, 224–226. [Google Scholar] [CrossRef]
- Li, Y.; Ye, D.; Chen, X.; Lu, X.; Shao, Z.; Zhang, H.; Che, Y. Breviane spiroditerpenoids from an extreme-tolerant Penicillium sp. isolated from a deep sea sediment sample. J. Nat. Prod. 2009, 72, 912–926. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ye, D.; Shao, Z.; Cui, C.; Che, Y. A sterol and spiroditerpenoids from a Penicillium sp. isolated from a deep sea sediment sample. Mar. Drugs 2012, 10, 497–508. [Google Scholar] [CrossRef]
- Li, D.; Wang, F.; Cai, S.; Zeng, X.; Xiao, X.; Gu, Q.; Zhu, W. Two new bisorbicillinoids isolated from a deep-sea fungus, Phialocephala sp. Fl30r. J. Antibiot. 2007, 60, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cai, S.; Zhu, T.; Wang, F.; Xiao, X.; Gu, Q. Three new sorbicillin trimers, trisorbicillinones B, C and D, from a deep ocean sediment derived fungus, Phialocephala sp. Fl30r. Tetrahedron 2010, 66, 5101–5106. [Google Scholar] [CrossRef]
- Li, D.-H.; Cai, S.-X.; Zhu, T.-J.; Wang, F.-P.; Xiao, X.; Gu, Q.-Q. New cytotoxic metabolites from a deep-sea-derived fungus, Phialocephala sp., strain FL30r. Chem. Biodiv. 2011, 8, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Liu, D.; Shao, Z.; Proksch, P.; Lin, W. Eutypellazines A–M, thiodiketopiperazine -type alkaloids from deep sea derived fungus Eutypella sp. MCCC 3A00281. RSC Adv. 2017, 7, 33580–33590. [Google Scholar] [CrossRef] [Green Version]
- Niu, S.; Liu, D.; Shao, Z.; Proksch, P.; Lin, W. Eutypellazines N–S, new thiodiketo-piperazines from a deep sea sediment derived fungus Eutypella sp. With anti-VRE activities. Tetrahedron Lett. 2017, 58, 3695–3699. [Google Scholar] [CrossRef]
- Bao, J.; Zhai, H.; Zhu, K.; Yu, J.-H.; Zhang, Y.; Wang, Y.; Jiang, C.-S.; Zhang, X.; Zhang, Y.; Zhang, H. Bioactive pyridone alkaloids from a deep-sea-derived fungus Arthrinium sp. UJNMF0008. Mar. Drugs 2018, 16, 174. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Zhang, X.; Nong, X.; Wang, J.; Qi, S. Brevianamides and mycophenolic acid derivatives from the deep-sea-derived fungus Penicillium brevicompactum DFFSCS025. Mar. Drugs 2017, 15, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredimoses, M.; Zhou, X.; Lin, X.; Tian, X.; Ai, W.; Wang, J.; Liao, S.; Liu, J.; Yang, B.; Yang, X.; et al. New prenylxanthones from the deep-sea derived fungus Emericella sp. SCSIO 05240. Mar. Drugs 2014, 12, 3190–3202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; He, W.; Huang, X.; Tian, X.; Liao, S.; Yang, B.; Wang, F.; Zhou, X.; Liu, Y. Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agric. Food Chem. 2016, 64, 2910–2916. [Google Scholar] [CrossRef]
- Wang, W.; Chen, R.; Luo, Z.; Wang, W.; Chen, J. Antimicrobial activity and molecular docking studies of a novel anthraquinone from a marine-derived fungus Aspergillus versicolor. Nat. Prod. Res. 2017, 32, 558–563. [Google Scholar] [CrossRef]
- Du, L.; Yang, X.; Zhu, T.; Wang, F.; Xiao, X.; Park, H.; Gu, Q. Diketopiperazine alkaloids from a deep ocean sdiment derived fungus Penicillium sp. Chem. Pharm. Bull. 2009, 57, 873–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, S.; Fan, Z.-W.; Xie, C.-L.; Liu, Q.; Luo, Z.-H.; Liu, G.; Yang, X.-W. Spirograterpene a, a tetracyclic spiro-diterpene with a fused 5/5/5/5 ring system from the deep-sea-derived fungus Penicillium granulatum MCCC 3A00475. J. Nat. Prod. 2017, 80, 2174–2177. [Google Scholar] [CrossRef]
- Bandaranayake, W.M. Traditional and medicinal uses of mangroves. Wetl. Ecol. Manag. Mangroves Salt Marshes 1998, 2, 133–148. [Google Scholar] [CrossRef]
- Thatoi, H.; Behera, B.C.; Mishra, R.R. Ecological role and biotechnological potential of mangrove fungi: A review. Mycology 2013, 4, 54–71. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Agrawal, S.; Prakash, V.; Gupta, M.K.; Reddy, M.S. Anti-infectives from mangrove endophytic fungi. S. A. J. Bot. 2020, 134, 237–263. [Google Scholar] [CrossRef]
- Cadamuro, R.D.; da Silveira Bastos, I.M.A.; Silva, I.T.; Cabral da Cruz, A.C.; Robl, D.; Sandjo, L.P.; Alves, S., Jr.; Lorenzo, J.M.; Rodríguez-Lázaro, D.; Treichel, H.; et al. Bioactive compounds from mangrove endophytic fungus and their uses for microorganism control. J. Fungi 2021, 7, 455. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wu, W.; Liu, X.; Zaleta-Pinet, D.A.; Clark, B.R. Bioactive compounds isolated from marine-derived microbes in China: 2009–2018. Mar. Drugs 2019, 17, 339. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xia, G.; Li, H.; Ma, L.; Ding, B.; Lu, Y.; He, L.; Xia, X.; She, Z. Vermistatin derivatives with α-glucosidase inhibitory activity from the mangrove endophytic fungus Penicillium sp. HN29-3B1. Planta Med. 2014, 80, 912–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yang, Q.; Xia, G.; Huang, H.; Li, H.; Ma, L.; Lu, Y.; He, L.; Xia, X.; She, Z. Polyketides with α-glucosidase inhibitory activity from a mangrove endophytic fungus, Penicillium sp. HN29-3B1. J. Nat. Prod. 2015, 78, 1816–1822. [Google Scholar] [CrossRef]
- Li, H.; Huang, H.; Shao, C.; Huang, H.; Jiang, J.; Zhu, X.; Liu, Y.; Liu, L.; Lu, Y.; Li, M.; et al. Cytotoxic norsesquiterpene peroxides from the endophytic fungus Talaromyces flavus isolated from the mangrove plant Sonneratia apetala. J. Nat. Prod. 2011, 74, 1230–1235. [Google Scholar] [CrossRef]
- Liu, H.; Chen, S.; Liu, W.; Liu, Y.; Huang, X.; She, Z. Polyketides with immunosuppressive activities from mangrove endophytic fungus Penicillium sp. ZJ-SY2. Mar. Drugs 2016, 14, 217. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.; Chen, S.; Long, Y.; Li, C.; Huang, X.; She, Z. Depsidones from Talaromyces stipitatus SK-4, an endophytic fungus of the mangrove plant Acanthus ilicifolius. Phytochem. Lett. 2017, 20, 196–199. [Google Scholar] [CrossRef]
- Nicoletti, R.; Salvatore, M.M.; Andolfi, A. Secondary metabolites of mangrove-associated strains of Talaromyces. Mar. Drugs 2018, 16, 12. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.J.; Zhu, T.J.; Fang, Y.C.; Gu, Q.Q.; Zhu, W.M. Polyketides from Penicillium sp JP-1, an endophytic fungus associated with the mangrove plant Aegiceras corniculatum. Phytochemistry 2008, 69, 1273–1278. [Google Scholar] [CrossRef]
- Xu, M.; Deng, Z.; Li, M.; Li, J.; Fu, H.; Proksch, P.; Lin, W. Chemical constituents from the mangrove plant, Aegiceras corniculatum. J. Nat. Prod. 2004, 67, 762–766. [Google Scholar] [CrossRef]
- Bandaranayake, W.M. Bioactivities, bioactive compounds and chemical constituents of mangrove plants. Wetlands Ecol. Managmt. 2002, 10, 421–452. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Ren, W.; Zhao, D.; Zhu, Y.; Wu, X. Bioactive metabolites from Chaetomium globosum l18, an endophytic fungus in the medicinal plant Curcuma wenyujin. Phytomedicine 2012, 19, 364–368. [Google Scholar] [CrossRef]
- Huang, S.; Chen, H.; Li, W.; Zhu, X.; Ding, W.; Li, C. Bioactive chaetoglobosins from the mangrove endophytic fungus Penicillium chrysogenum. Mar. Drugs 2016, 14, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Gong, B.; Cox, D.G.; Li, C.; Wang, J.; Ding, W. Dichlorodiaportinol a—A new chlorine-containing isocoumarin from an endophytic fungus Trichoderma sp. 09 from Myoporum bontioides a. Gray and its cytotoxic activity. Pharmacogn. Mag. 2014, 10, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Xu, J.; Li, F.; Xu, L.; Li, C. A new antifungal isocoumarin from the endophytic fungus Trichoderma sp. 09 of Myoporum bontioides a. Gray. Pharmacogn. Mag. 2016, 12, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiong, P.; Zheng, W.; Zhu, X.; She, Z.; Ding, W.; Li, C. Identification and antifungal activity of compounds from the mangrove endophytic fungus Aspergillus clavatus R7. Mar. Drugs 2017, 15, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, G.; Zhu, X.; Li, Q.; Gu, M.; He, Z.; Wu, J.; Li, J.; Lin, Y.; Li, M.; She, Z.; et al. SZ-685C, a marine anthraquinone, is a potent inducer of apoptosis with anticancer activity by suppression of the Akt/FOXO pathway. Br. J. Pharmacol. 2010, 159, 689–697. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Li, M.; Jun, L.; Li, M.; Li, W.; Lin, Y.; Liu, H.; She, Z.; Wu, J.; Xia, J.; et al. Application of Anthracycline Compound in Preparing Anti-Breast Cancer Medicines. Patent Number CN101862313, 17 May 2010. [Google Scholar]
- Zhu, X.; He, Z.; Wu, J.; Yuan, J.; Wen, W.; Hu, Y.; Jiang, Y.; Lin, C.; Zhang, Q.; Lin, M.; et al. A marine anthraquinone SZ-685C overrides adriamycin-resistance in breast cancer cells through suppressing Akt signaling. Mar. Drugs 2012, 10, 694–711. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-H.; Xiao, W.-W.; Jiang, X.-B.; Wang, J.-W.; Mao, Z.-G.; Lei, N.; Fan, X.; Song, B.-B.; Liao, C.-X.; Wang, H.-J.; et al. A novel marine drug, SZ-685C, induces apoptosis of MMQ pituitary tumor cells by downregulating miR-200c. Curr. Med. Chem. 2013, 20, 2145–2154. [Google Scholar] [CrossRef]
- Wang, D.; Wang, S.; Liu, Q.; Wang, M.; Wang, C.; Yang, H. SZ-685C exhibits potent anticancer activity in both radiosensitive and radioresistant npc cells through the miR-205-PTEN-Akt pathway. Oncol. Rep. 2013, 29, 2341–2347. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Tan, T.; Mao, Z.-G.; Lei, N.; Wang, Z.-Y.; Hu, B.; Chen, Z.-Y.; She, Z.-G.; Zhu, Y.-H.; Wang, H.-J. The marine metabolite SZ-685C induces apoptosis in primary human nnfunctioning pituitary adenoma cells by inhibition of the Akt pathway in vitro. Mar. Drugs 2015, 13, 1569–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenical, W.; Jensen, P.R.; Kauffman, C.; Mayhead, S.L.; Faulkner, D.J.; Sincich, C.; Rao, M.R.; Kantorowski, E.J.; West, L.M.; Strangman, W.K.; et al. New anticancer drugs from cultured and collected marine organisms. Pharm. Biol. 2003, 41, 6–14. [Google Scholar] [CrossRef] [Green Version]
- Gullo, V.P.; McAlpine, J.; Lam, K.S.; Baker, D.; Petersen, F. Drug discovery from natural products. J. Ind. Microbiol. Biotechnol. 2006, 33, 523–531. [Google Scholar] [CrossRef]
- BeyondSpringPharma. Pliabulin and Docetaxel. Available online: https://bit.ly/3inGCfV (accessed on 1 December 2021).
- Kim, D.-H.; Brunt, J.; Austin, B. Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhynchus mykiss). J. Appl. Microbiol. 2006, 102, 1654–1664. [Google Scholar] [CrossRef]
- Ward, N.L.; Steven, B.; Penn, K.; Methe, B.A.; Detrich III, W.H. Characterization of the intestinal microbiota of two antarctic notothenioid fish species. Extremophiles 2009, 13, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Smriga, S.; Sandin, S.A.; Azam, F. Abundance, diversity, and activity of microbial assemblages associated with coral reef fish guts and feces. FEMS Microbiol. Ecol. 2010, 73, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595–1608. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, L.M.; Wong, W.R.; Riener, R.M.; Schulze, C.J.; Linington, R.G. Examining the fish microbiome: Vertebrate-derived bacteria as an environmental niche for the discovery of unique marine natural products. PLoS ONE 2012, 7, e35398. [Google Scholar] [CrossRef] [PubMed]
- Numata, A.; Takahashi, C.; Matsushita, T.; Miyamoto, T.; Kawai, K.; Usami, Y.; Matsumura, E.; Inoue, M.; Ohishi, H.; Shingu, T. Fumiquinazolines, novel metabolites of a fungus isolated from a saltfish. Tetrahedron Lett. 1992, 33, 1621–1624. [Google Scholar] [CrossRef]
- Takahashi, C.; Matsushita, T.; Doi, M.; Minoura, K.; Shingu, T.; Kumeda, Y.; Numata, A. Fumiquinazolines A-G, novel metabolites of a fungus separated from a Pseudolabrus marine fish. J. Chem. Soc. Perkin Trans. 1995, 1, 2345–2353. [Google Scholar] [CrossRef]
- Belofsky, G.N.; Anguera, M.; Jensen, P.R.; Fenical, W.; Kock, M. Oxepinamides A-C and fumiquinazolines H-I: Bioactive metabolites from a marine isolate of a fungus of the genus Acremonium. Chem.-Eur. J. 2000, 6, 1355–1360. [Google Scholar] [CrossRef]
- Shao, C.L.; Xu, R.F.; Wei, M.Y.; She, Z.G.; Wang, C.Y. Structure and absolute configuration of fumiquinazoline l, an alkaloid from a gorgonian-derived Scopulariopsis sp. fungus. J. Nat. Prod. 2013, 76, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Resende, D.I.S.P.; Boonpothong, P.; Sousa, E.; Kijjoa, A.; Pinto, M.M.M. Chemistry of the fumiquinazolines and structurally related alkaloids. Nat. Prod. Rep. 2019, 36, 7–34. [Google Scholar] [CrossRef]
- Han, J.; Liu, M.; Jenkins, I.D.; Liu, X.; Zhang, L.; Quinn, R.J.; Feng, Y. Genome-inspired chemical exploration of marine fungus Aspergillus fumigatus MF071. Mar. Drugs 2020, 18, 352. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Doi, M.; Shigeta, H.; Muroga, Y.; Hosoe, S.; Numata, A.; Tanaka, R. Absolute stereostructures of cytotoxic metabolites, chaetomugilins A-C, produced by a Chaetomium species separated from a marine fish. Tetrahedron Lett. 2008, 49, 4192–4195. [Google Scholar] [CrossRef]
- Yasuhide, M.; Yamada, T.; Numata, A.; Tanaka, R. Chaetomugilins, new selectively cytotoxic metabolites, produced by a marine fish-derived Chaetomium species. J. Antibiot. 2008, 61, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Yasuhide, M.; Shigeta, H.; Numata, A.; Tanaka, R. Absolute stereostructures of chaetomugilins G and H produced by a marine-fish-derived Chaetomium species. J. Antibiot. 2009, 62, 353–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muroga, Y.; Yamada, T.; Numata, A.; Tanaka, R. Chaetomugilins I-O, new potent cytotoxic metabolites from a marine-fish-derived Chaetomium species. Stereochemistry and biological activities. Tetrahedron 2009, 65, 7580–7586. [Google Scholar] [CrossRef]
- Hu, X.; Wang, J.; Chai, J.; Yu, X.; Zhang, Y.; Feng, Y.; Qin, J.; Yu, H. Chaetomugilin J enhances apoptosis in human ovarian cancer A2780 cells induced by cisplatin through inhibiting pink1/parkin mediated mitophagy. Oncotargets Therap. 2020, 13, 9967–9976. [Google Scholar] [CrossRef]
- Yamada, T.; Muroga, Y.; Tanaka, R. New azaphilones, seco-chaetomugilins A and D, produced by a marine-fish-derived Chaetomium globosum. Mar. Drugs 2009, 7, 249–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muroga, Y.; Yamada, T.; Numata, A.; Tanaka, R. 11- and 4’-epimers of chaetomugilin A, novel cytostatic metabolites from marine fish-derived fungus Chaetomium globosum. Helv. Chim. Acta 2010, 93, 542–549. [Google Scholar] [CrossRef]
- Winter, J.M.; Sato, M.; Sugimoto, S.; Chiou, G.; Garg, N.K.; Tang, Y.; Watanabe, K. Identification and characterization of the chaetoviridin and chaetomugilin gene cluster in Chaetomium globosum reveal dual functions of an iterative highly-reducing polyketide synthase. J. Am. Chem. Soc. 2012, 134, 17900–17903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prompanya, C.; Dethoup, T.; Bessa, L.J.; Pinto, M.M.M.; Gales, L.; Costa, P.M.; Silva, A.M.S.; Kijjoa, A. New isocoumarin derivatives and meroterpenoids from the marine sponge-associated fungus aspergillus similanensis sp. Nov. Kufa 0013. Mar. Drugs 2014, 12, 5160–5173. [Google Scholar] [CrossRef]
- Prompanya, C.; Fernandes, C.; Cravo, S.; Pinto, M.M.M.; Dethoup, T.; Silva, A.M.S.; Kijjoa, A. A new cyclic hexapeptide and a new isocoumarin derivative from the marine sponge-associated fungus Aspergillus similanensis KUFA 0013. Mar. Drugs 2015, 13, 1432–1450. [Google Scholar] [CrossRef] [Green Version]
- Masuda, Y.; Tanaka, R.; Ganesan, A.; Doi, D. Structure revision of similanamide to PF1171C by total synthesis. J. Nat. Prod. 2015, 78, 2286–2291. [Google Scholar] [CrossRef]
- Kai, K.; Yoshikawa, H.; Kuo, Y.-H.; Akiyama, K.; Haysahi, H. Determination of absolute structures of cyclic peptides, PF1171a and PF1171c, from unidentified Ascomycete OK-128. Biosci. Biotech. Biochem. 2010, 74, 1309–1311. [Google Scholar] [CrossRef]
- Pang, X.; Lin, X.; Wang, P.; Zhou, X.; Yang, B.; Wang, J.; Liu, Y. Perylenequione derivatives with anticancer activities isolated from the marine sponge-derived fungus, Alternaria sp. SCSIO41014. Mar. Drugs 2018, 16, 280. [Google Scholar] [CrossRef] [Green Version]
- Ur Rehman, S.; Yang, L.-J.; Zhang, Y.-H.; Wu, J.-S.; Shi, T.; Haider, W.; Shao, C.-L.; Wang, C.-Y. Sorbicillinoid derivatives from sponge-derived fungus Trichoderma reesei (HN-2016-018). Front. Microbiol. 2020, 11, 1334. [Google Scholar] [CrossRef]
- Jiang, W.; Ye, P.; Chen, C.T.A.; Wang, K.; Liu, P.; He, S.; Wu, X.; Gan, L.; Ye, Y.; Wu, B. Two novel hepatocellular carcinoma cycle inhibitory cyclodepsipeptides from a hydrothermal vent crab-associated fungus Aspergillus clavatus C2WU. Mar. Drugs 2013, 11, 4761–4772. [Google Scholar] [CrossRef]
- Takahashi, K.; Sakai, K.; Nagano, Y.; Sakaguchi, S.O.; Lima, A.O.; Pellizari, V.H.; Iwatsuki, M.; Takishita, K.; Nonaka, K.; Fujikura, K.; et al. Cladomarine, a new anti-saprolegniasis compound isolated from the deep-sea fungus, Penicillium coralligerum YK-247. J. Antibiot. 2017, 70, 911–914. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, Y.-Y.; Shao, C.-L.; Wang, C.-Y. Metabolites from marine invertebrates and their symbiotic microorganisms: Molecular diversity discovery, mining, and application. Mar. Life Sci. Technol. 2019, 1, 60–94. [Google Scholar] [CrossRef] [Green Version]
- Marchese, P.; Young, R.; O’Connell, E.; Afoullouss, S.; Baker, B.J.; Allcock, A.L.; Barry, F.; Murphy, J.M. Deep-sea coral garden invertebrates and their associated fungi are genetic resources for chronic disease drug discovery. Mar. Drugs 2021, 19, 390. [Google Scholar] [CrossRef]
- Rust, M.; Helfrich, E.J.N.; Freeman, M.F.; Nanudorn, P.; Field, C.M.; Rückert, C.; Kündig, T.; Page, M.J.; Webb, V.L.; Kalinowski, J.; et al. A multiproducer microbiome generates chemical diversity in the marine sponge Mycale hentscheli. Proc. Natl. Acad. Sci. USA 2020, 117, 9508–9518. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.C.; Mori, T.; Ruckert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.-W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, Y.-M.; Szewczyk, E.; Nayak, T.; Davidson, A.D.; Sanchez, J.F.; Lo, H.-C.; Ho, W.-H.; Simityan, H.; Kuo, E.; Praseuth, A.; et al. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem. Biol. 2008, 15, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.X.; Ding, L.; He, S. Discovery of a new biphenyl derivative by epigenetic manipulation of marine-derived fungus Aspergillus versicolor. Nat. Prod. Res. 2019, 33, 1191–1195. [Google Scholar] [CrossRef]
- He, X.; Zhang, Z.; Chen, Y.; Che, Q.; Zhu, T.; Gu, Q.; Li, D. Varitatin A, a highly modified fatty acid amide from Penicillium variabile cultured with a DNA methyltransferase inhibitor. J. Nat. Prod. 2015, 78, 2841–2845. [Google Scholar] [CrossRef]
- He, X.; Zhang, Z.; Che, Q.; Zhu, T.; Gu, Q.; Li, D. Varilactones and wortmannilactones produced by Penicillium variabile cultured with histone deacetylase inhibitor. Arch. Pharmacal Res. 2018, 41, 57–63. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Tang, J.; Li, X. Eremophilane sesquiterpenes from a deep marine-derived fungus, Aspergillus sp. SCSIOW2, cultivated in the presence of epigenetic modifying agents. Molecules 2016, 21, 473. [Google Scholar] [CrossRef] [Green Version]
- Beau, J.; Mahid, N.; Burda, W.N.; Harrington, L.; Shaw, L.N.; Mutka, T.; Kyle, D.E.; Barisic, B.; van Olphen, A.; Baker, B.J. Epigenetic tailoring for the production of anti-iinfective cytosporones from the marine fungus Leucostoma persoonii. Mar. Drugs 2012, 10, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shao, C.-L.; Chen, M.; Liu, Q.-A.; Wang, C.-Y. Brominated resorcylic acid lactones from the marine-derived fungus Cochliobolus lunatus induced by histone deacetylase inhibitors. Tetrahedron Lett. 2014, 55, 4888–4891. [Google Scholar] [CrossRef]
- Zahner, H. What are secondary metabolites? Folia Microbiol. 1979, 24, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Bu’Lock, J.D.; Nisbet, L.J.; Winstanley, D.J. Bioactive Microbial Products: Search and Discovery; Academic Press: London, UK, 1982; Zahner Chapter; p. 148. ISBN 0-12-140750-0. [Google Scholar] [CrossRef]
- Chiang, Y.-M.; Lee, K.-H.; Sanchez, J.F.; Keller, N.P.; Wang, C.C.C. Unlocking fungal cryptic natural products. Nat. Prod. Comm. 2009, 4, 1505–1510. [Google Scholar] [CrossRef] [Green Version]
- Hemphill, C.F.P.; Sureechatchaiyan, P.; Kassack, M.U.; Orfali, R.S.; Lin, W.; Daletos, G.; Proksch, P. OSMAC approach leads to new fusarielin metabolites from Fusarium tricinctum. J. Antibiot. 2017, 70, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhu, T.; Wei, H.; Zhang, G.; Wang, H.; Gu, Q. Spicochalasin A and new aspochalasins from the marine-derived fungus Spicaria elegans. Eur. J. Org. Chem. 2009, 3045–3051. [Google Scholar] [CrossRef]
- Wu, H.; Ding, Y.; Hu, K.; Long, X.; Qu, C.; Puno, P.-T.; Deng, J. Bioinspired network analysis enabled divergent syntheses and structure revision of pentacyclic cytochalasans. Angew. Chem. Int. Ed. 2021, 60, 15963–15971. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Gu, Q.; Zhu, W.; Cui, C.; Fan, G.; Fang, Y.; Zhu, T.; Liu, H. 10-phenyl-[12]-cytochalasins Z7, Z8, and Z9 from the marine-derived fungus Spicaria elegans. J. Nat. Prod. 2006, 69, 871–875. [Google Scholar] [CrossRef]
- Liu, R.; Lin, Z.; Zhu, T.; Fang, Y.; Gu, Q.; Zhu, W. Novel open-chain cytochalasins from the marine-derived fungus Spicaria elegans. J. Nat. Prod. 2008, 71, 1127–1132. [Google Scholar] [CrossRef]
- Shang, Z.; Li, X.-M.; Li, C.-S.; Wang, B.-G. Diverse secondary metabolites produced by marine-derived fungus Nigrospora sp. MA75 on various culture media. Chem. Biodiv. 2012, 9, 1338–1348. [Google Scholar] [CrossRef]
- Huang, H.; Wang, F.; Luo, M.; Chen, Y.; Song, Y.; Zhang, W.; Zhang, S.; Ju, J. Halogenated anthraquinones from the marine-derived fungus Aspergillus sp. SCSIO F063. J. Nat. Prod. 2012, 75, 1346–1352. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Z.; Sun, K.; Zhu, W. Effects of high salt stress on secondary metabolite production in the marine-derived fungus Spicaria elegans. Mar. Drugs 2011, 9, 535–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, Y.; Wei, H.; Zhang, Z.; Che, Q.; Liu, Y.; Zhu, T.; Mándi, A.; Kurtán, T.; Gu, Q.; Li, D. Eleganketal A, a highly oxygenated dibenzospiroketal from the marine-derived fungus Spicaria elegans KLA03. J. Nat. Prod. 2014, 77, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Yun, K.; Feng, Z.; Choi, H.D.; Kang, J.S.; Son, B.W. New production of (R)-(−)-5-bromomellein, a dihydroisocoumarin derivative from the marine-derived fungus Aspergillus ochraceus. Chem. Nat. Compd. 2013, 49, 24–26. [Google Scholar] [CrossRef]
- Wang, F.-Z.; Wei, H.-J.; Zhu, T.-J.; Li, D.-H.; Lin, Z.-J.; Gu, Q.-Q. Three new cytochalasins from the marine-derived fungus Spicaria elegans KLA03 by supplementing the cultures with L- and D-tryptophan. Chem. Biodiv. 2011, 8, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-X.; Xu, M.-Y.; Li, H.J.; Zeng, K.-J.; Ma, W.-Z.; Tian, G.-B.; Xu, J.; Yang, D.-P.; Lan, W.-J. Diverse secondary metabolites from the marine-derived fungus Dichotomomyces cejpii F31-1. Mar. Drugs 2017, 15, 339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Zhong, Y.; Shen, L.; Wu, X.; Ye, Y.; Chen, C.-T.A.; Wu, B. Stress-driven discovery of natural products from extreme marine environment-Kueishantao hydrothermal vent, a case study of metal switch valve. Curr. Org. Chem. 2014, 18, 925–934. [Google Scholar] [CrossRef]
- Ye, P.; Shen, L.; Jiang, W.; Ye, Y.; Chen, C.T.; Wu, X.; Wang, K.; Wu, B. Zn-driven discovery of a hydrothermal vent fungal metabolite clavatustide C, and an experimental study of the anti-cancer mechanism of clavatustide B. Mar. Drugs 2014, 12, 3203–3217. [Google Scholar] [CrossRef] [Green Version]
- Ding, C.; Wu, X.; Auckloo, B.N.; Chen, C.-T.A.; Ye, Y.; Wang, K.; Wu, B. An unusual stress metabolite from a hydrothermal vent fungus Aspergillus sp. WU 243 induced by cobalt. Molecules 2016, 21, 105. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-J.; Li, D.-Y.; Li, Y.-C.; Hua, H.-M.; Ma, E.-L.; Li, Z.-L. Caryophyllene sesquiterpenes from the marine-derived fungus Ascotricha sp. ZJ-M-5 by the one strain–many compounds strategy. J. Nat. Prod. 2014, 77, 1367–1371. [Google Scholar] [CrossRef]
- Chen, X.-W.; Li, C.-W.; Cui, C.B.; Hua, W.; Zhu, T.-J.; Gu, Q.-Q. Nine new and five known polyketides derived from a deep sea-sourced Aspergillus sp. 16-02-1. Mar. Drugs 2014, 12, 3116–3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.W.; Li, C.W.; Hua, W.; Wu, C.J.; Cui, C.B.; Zhu, T.J.; Gu, Q.Q. Metabolites of Aspergillus sp. 16-02-1 isolated from a deep sea sediment and preliminary test of their antitumor and antifungal activities. Chin. J. Mar. Drugs 2013, 32, 1–10. [Google Scholar]
- Guo, W.; Zhang, Z.; Zhu, T.; Gu, Q.; Li, D. Penicyclones A-E, antibacterial polyketides from the deep-sea-derived fungus Penicillium sp. F23-2. J. Nat. Prod. 2015, 78, 2699–2703. [Google Scholar] [CrossRef]
- Wu, B.; Wiese, J.; Labes, A.; Kramer, A.; Schmaljohann, R.; Imhoff, J.F. Lindgomycin, an unusual antibiotic polyketide from a marine fungus of the Lindgomycetaceae. Mar. Drugs 2015, 13, 4617–4632. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.-S.; Rong, X.-G.; Kang, H.-H.; Ma, L.-Y.; Hamann, M.; Liu, W.-Z. Raistrickiones A–E from a highly productive strain of Penicillium raistrickii generated through thermo change. Mar. Drugs 2018, 16, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christian, O.E.; Compton, J.; Christian, K.R.; Mooberry, S.L.; Valeriote, F.A.; Crews, P. Using jasplakinolide to turn on pathways that enable the isolation of new chaetoglobosins from Phomospis asparagi. J. Nat. Prod. 2005, 68, 1592–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.-J.; Zhu, T.-J.; Zhang, G.-J.; Wei, H.-J.; Gu, Q.-Q. Deoxycytochalasins from a marine-derived fungus Spicaria elegans. Can. J. Chem. 2009, 87, 486–489. [Google Scholar] [CrossRef]
- Reen, J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The sound of silence: Activating silent biosynthetic gene cllusters in marine microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.D.; Savard, M.E. Antibiotic activity of the marine fungus Leptosphaeria oraemaris. Proc. N. S. Inst. Sci. 1989, 39, 51–58. Available online: https://dalspace.library.dalca//handle/10222/35250 (accessed on 12 December 2021).
- Masuma, R.; Yamaguchi, Y.; Noumi, M.; Omura, S.; Namikoshi, M. Effect of sea water concentration on hyphal growth and antimicrobial metabolite production in marine fungi. Mycoscience 2001, 42, 455–459. [Google Scholar] [CrossRef]
- Mikolasch, A.; Hessel, S.; Salazar, M.G.; Neumann, H.; Manda, K.; Gōrdes, D.; Schmidt, E.; Thurow, K.; Hammer, E.; Lindequist, U.; et al. Synthesis of new N-analogous corollosporine derivatives with antibacterial activity by laccase-catalyzed amination. Chem. Pharm. Bull. 2008, 56, 781–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roullier, C.; Bertrand, S.; Blanchet, E.; Peigné, M.; Robiou du Pont, T.; Guitton, Y.; Pouchus, Y.F.; Grovel, O. Time dependency of chemodiversity and biosynthetic pathways: An LC-MS metabolomic study of marine-sourced Penicillium. Mar. Drugs 2016, 14, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbanat, D.; Leighton, M.; Maiese, W.; Jones, E.B.; Pearce, C.; Greenstein, M. Cell wall active antifungal compounds produced by the marine fungus Hypoxylon oceanicum LL-15G256. I. Taxonomy and fermentation. J. Antibiot. 1998, 51, 296–302. [Google Scholar] [CrossRef]
- Doshida, J.; Hasegawa, H.; Onuki, H.; Shimidzu, N. Exophilin A, a new antibiotic from a marine microorganism Exophiala pisciphila. J. Antibiot. 1996, 49, 1105–1109. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Yin, Y.; Zhang, F.; Li, Z.; Wang, L. Operating conditions optimization for (+)-terrein production in a stirred bioreactor by Aspergillus terreus strain PF-26 from marine sponge Phakellia fusca. Bioproc. Biosys. Engin. 2012, 35, 1651–1655. [Google Scholar] [CrossRef]
- Pignatiello, J.J., Jr. An overview of the strategy and tactics of Tagaguchi. IEEE Trans. 1988, 20, 247–254. [Google Scholar] [CrossRef]
- Plackett, R.L.; Burman, J.P. The design of optimum multifactorial experiments. Biometrika 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Cochran, W.G.; Cox, G.M. Experimental Designs; John Wiley and Sons: New York, NY, USA, 1957; p. 611. ISBN 9780471162049. [Google Scholar]
- Du, L.; Zhu, T.J.; Fang, Y.C.; Liu, H.B.; Gu, Q.Q.; Zhu, W.M. Aspergiolide Aone, a novel anthraquinone derivative with naphtho[1,2,3-de]chromene-2,7-dione skeleton isolated from a marine-derived fungus Aspergillus glaucus. Tetrahedron 2007, 63, 1085–1088. [Google Scholar] [CrossRef]
- Cai, M.-H.; Zhou, X.-S.; Sun, X.-Q.; Tao, K.-J.; Zhang, Y.-X. Statistical optimization of medium composition for aspergiolide A production by marine-derived fungus Aspergillus glaucus. J. Indust. Microbiol. Biotech. 2009, 36, 381–389. [Google Scholar] [CrossRef]
- Zhou, S.-l.; Wang, M.; Zhao, H.-S.; Huang, Y.-h.; Lin, Y.-y.; Tan, G.-h.; Chen, S.-l. Penicilazaphilone C, a new antineoplastic and antibacterial azaphilone from the marine fungus Penicillium sclerotiorum. Archiv. Pharmacal Res. 2016, 39, 1621–1627. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-G.; Wang, M.; Lin, Y.-Y.; Zhou, S.-L. Optimization of culture conditions for penicilazaphilone C production by a marine-derived fungus Penicillium sclerotiorum M-22H. Lett. Appl. Microbiol. 2017, 66, 222–230. [Google Scholar] [CrossRef]
- Yu, Z.; Lang, G.; Kajahn, I.; Schmaljohann, R.; Imhoff, J.F. Scopularides A and B, cyclodepsipeptides from a marine sponge-derived fungus, Scopulariopsis brevicaulis. J. Nat. Prod. 2008, 71, 1052–1054. [Google Scholar] [CrossRef]
- Ding, L.-J.; Yuan, W.; Liao, X.-J.; Han, B.-N.; Wang, S.-P.; Li, Z.-Y.; Xu, S.-H.; Zhang, W.; Lin, H.-W. Oryzamides A-E, cyclodepsipeptides from the sponge-derived fungus Nigrospora oryzae PF18. J. Nat. Prod. 2016, 79, 2045–2052. [Google Scholar] [CrossRef]
- Kramer, A.; Paun, L.; Imhoff, J.F.; Kempken, F.; Labes, A. Development and validation of a fast and optimized screening method for enhanced production ofsecondary metabolites using the marine Scopulariopsis brevicaulis strain LF580 producing anti-cancer active scopularide A and B. PLoS ONE 2014, 9, e103320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, A.; Beck, H.C.; Kumar, A.; Kristensen, L.P.; Imhoff, J.F.; Labes, A. Proteomic analysis of anti-cancerous scopularide production by a marine Microascus brevicaulis strain and its UV mutant. PLoS ONE 2015, 10, e0140047. [Google Scholar] [CrossRef] [PubMed]
- Lukassen, M.B.; Saei, W.; Sondergaard, T.E.; Tamminen, A.; Kumar, A.; Kempken, F.; Wiebe, M.G.; Sørensen, J.L. Identification of the scopularide biosynthetic gene cluster in Scopulariopsis brevicaulis. Mar. Drugs 2015, 13, 4331–4343. [Google Scholar] [CrossRef] [Green Version]
- Alberti, F.; Foster, G.D.; Bailey, A.M. Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol. 2017, 101, 493–500. [Google Scholar] [CrossRef] [Green Version]
- Boecker, S.; Grätz, S.; Kerwat, D.; Adam, L.; Schirmer, D.; Richter, L.; Schütze, T.; Petras, D.; Süssmuth, R.D.; Meyer, V. Aspergillus niger is a superior expression host for the production of bioactive fungal cyclodepsipeptides. Fungal Biol. Biotech. 2018, 5, 4. [Google Scholar] [CrossRef]
- Chaichanan, J.; Wiyakrutta, S.; Pongtharangkul, T.; Isarangkul, D.; Meevootisom, V. Optimization of zofimarin production by an endophytic fungus, Xylaria sp. Acra L38. Braz. J. Microbiol. 2014, 45, 287–293. [Google Scholar] [CrossRef] [Green Version]
- Reich, M.; Labes, A. How to boost marine fungal research: A first step towards a multidisciplinary approach by combining molecular fungal ecology and natural products chemistry. Mar. Gent. 2017, 36, 57–75. [Google Scholar] [CrossRef]
- Nai, C.; Meyer, V. From axenic to mixed cultures: Technological advances accelerating a paradigm shift in microbiology. Trends Microbiol. 2018, 26, 538–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stierle, A.A.; Stierle, D.B.; Decato, D.; Priestley, N.D.; Alverson, J.B.; Hoody, J.; McGrath, K.; Klepacki, D. The berkeleylactones, antibiotic macrolides from fungal coculture. J. Nat. Prod. 2017, 80, 1150–1160. [Google Scholar] [CrossRef] [Green Version]
- Nie, Y.; Yang, W.; Liu, Y.; Yang, J.; Lei, X.; Gerwick, W.H.; Zhang, Y. Acetylcholinesterase inhibitors and antioxidants mining from marine fungi: Bioassays, bioactivity coupled LC–MS/MS analyses and molecular networking. Mar. Life Sci. Technol. 2020, 2, 386–397. [Google Scholar] [CrossRef]
- Lage, O.M.; Ramos, M.C.; Calisto, R.; Almeida, E.; Vasconcelos, V.; Vicente, F. Current screening methodologies in drug discovery for selected human diseases. Mar. Drugs 2018, 16, 279. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Zheng, G.; Lu, Y. Recent advances in strategies for the cloning of natural product biosynthetic gene clusters. Front. Bioeng. Biotechnol. 2021, 9, 692797. [Google Scholar] [CrossRef] [PubMed]












Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giddings, L.-A.; Newman, D.J. Extremophilic Fungi from Marine Environments: Underexplored Sources of Antitumor, Anti-Infective and Other Biologically Active Agents. Mar. Drugs 2022, 20, 62. https://doi.org/10.3390/md20010062
Giddings L-A, Newman DJ. Extremophilic Fungi from Marine Environments: Underexplored Sources of Antitumor, Anti-Infective and Other Biologically Active Agents. Marine Drugs. 2022; 20(1):62. https://doi.org/10.3390/md20010062
Chicago/Turabian StyleGiddings, Lesley-Ann, and David J. Newman. 2022. "Extremophilic Fungi from Marine Environments: Underexplored Sources of Antitumor, Anti-Infective and Other Biologically Active Agents" Marine Drugs 20, no. 1: 62. https://doi.org/10.3390/md20010062
APA StyleGiddings, L. -A., & Newman, D. J. (2022). Extremophilic Fungi from Marine Environments: Underexplored Sources of Antitumor, Anti-Infective and Other Biologically Active Agents. Marine Drugs, 20(1), 62. https://doi.org/10.3390/md20010062