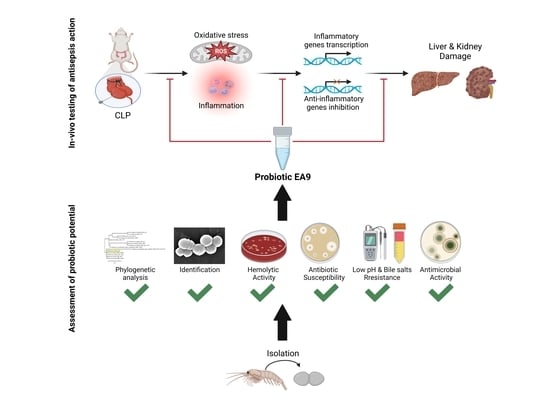
Probiotic Potential of the Marine Isolate Enterococcus faecium EA9 and In Vivo Evaluation of Its Antisepsis Action in Rats
Abstract
:1. Introduction
2. Results
2.1. Assessment of Probiotic Potential of Isolate EA9
2.1.1. Phenotypic and Biochemical Characterization of Isolate EA9
2.1.2. Molecular Identification
2.1.3. Blood Hemolysis
2.1.4. Antibiotic Resistance
2.1.5. Resistance to pH
2.1.6. Bile Salts Tolerance
2.1.7. Antimicrobial Activity
2.2. In Vivo Testing of the Antisepsis Action of the Isolated EA9 Probiotic
2.2.1. EA9 Effects on Liver and Kidney Functions Tests
2.2.2. EA9 Effects on Liver and Kidney Inflammation
2.2.3. EA9 Effects on Liver and Kidney Oxidative Stress
2.2.4. EA9 Effects on Liver and Kidney Histology
2.2.5. EA9 Effects on Liver and Kidney Gene Expressions
3. Discussion
4. Materials and Methods
4.1. Assessment of Probiotic Potential of Isolate EA9
4.1.1. Phenotypic and Biochemical Characterization of Isolate EA9
4.1.2. Molecular Identification and Phylogenetic Analysis of Isolate EA9
4.1.3. Hemolytic Activity
4.1.4. Antibiotic Susceptibility
4.1.5. Resistance to Low pH
4.1.6. Bile Salts Tolerance
4.1.7. Antimicrobial Activity
4.2. In Vivo Testing of the Antisepsis Action of the Isolated EA9 Probiotic
4.2.1. Ethical Approval and Experimental Model
4.2.2. Serum Biochemistry
4.2.3. Evaluation of Inflammation
4.2.4. Evaluation of Oxidative Damage
4.2.5. Histological Evaluation
4.2.6. Polymerase Chain Reaction (PCR) Assessment of Genes Expressions
4.2.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Florou-Paneri, P.; Christaki, E.; Bonos, E. Lactic Acid Bacteria as Source of Functional Ingredients. In Lactic Acid Bacteria-R & D for Food, Health and Livestock Purposes; Kongo, M., Ed.; IntechOpen: London, UK, 2013. [Google Scholar]
- Ou, Y.; Chen, S.; Ren, F.; Zhang, M.; Ge, S.; Guo, H.; Zhang, H.; Zhao, L. Lactobacillus casei Strain Shirota Alleviates Constipation in Adults by Increasing the Pipecolinic Acid Level in the Gut. Front. Microbiol. 2019, 10, 324. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-J.; Li, S.-H.; Li, A.-L.; Zhang, Q.-M.; Ni, W.-W.; Li, M.-N.; Meng, X.-C.; Li, C.; Jiang, S.-L.; Pan, J.-C.; et al. Effect of Lactobacillus acidophilus KLDS 1.0738 on miRNA expression in in vitro and in vivo models of β-lactoglobulin allergy. Biosci. Biotechnol. Biochem. 2018, 82, 1955–1963. [Google Scholar] [CrossRef] [PubMed]
- Fooladi, A.A.I.; Hosseini, H.M.; Nourani, M.R.; Khani, S.; Alavian, S.M. Probiotic as a Novel Treatment Strategy Against Liver Disease. Hepat. Mon. 2013, 13, e7521. [Google Scholar] [CrossRef] [Green Version]
- Mokoena, M.P.; Mutanda, T.; Olaniran, A.O. Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages. Food Nutr. Res. 2016, 60, 29630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, H.; Salminen, S.; Verhagen, H.; Rowland, I.; Heimbach, J.; Bañares, S.; Young, T.; Nomoto, K.; Lalonde, M. Novel probiotics and prebiotics: Road to the market. Curr. Opin. Biotechnol. 2015, 32, 99–103. [Google Scholar] [CrossRef]
- Román, L.; Real, F.; Sorroza, L.; Padilla, D.; Acosta, B.; Grasso, V.; Bravo, J.; Acosta, F. The in vitro effect of probiotic Vagococcus fluvialis on the innate immune parameters of Sparus aurata and Dicentrarchus labrax. Fish Shellfish. Immunol. 2012, 33, 1071–1075. [Google Scholar] [CrossRef]
- Sorroza, L.; Padilla, D.; Acosta, F.; Román, L.; Grasso, V.; Vega, J.; Real, F. Characterization of the probiotic strain Vagococcus fluvialis in the protection of European sea bass (Dicentrarchus labrax) against vibriosis by Vibrio anguillarum. Vet. Microbiol. 2012, 155, 369–373. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Yakovenko, E.P.; Strokova, T.V.; Ivanov, A.N.; Iakovenko, A.V.; Gioeva, I.Z.; Aldiyarova, M.A. The effectiveness of a probiotic containing Bifidobacterium longum BB-46 and Enterococcus faecium ENCfa-68 in the treatment of post-infectious irritable bowel syndrome. Prospective randomized comparative study. Ter. arkhiv 2022, 94, 180–187. [Google Scholar] [CrossRef]
- Ghazisaeedi, F.; Meens, J.; Hansche, B.; Maurischat, S.; Schwerk, P.; Goethe, R.; Wieler, L.H.; Fulde, M.; Tedin, K. A virulence factor as a therapeutic: The probiotic Enterococcus faecium SF68 arginine deiminase inhibits innate immune signaling pathways. Gut Microbes 2022, 14, 2106105. [Google Scholar] [CrossRef]
- Wu, Y.; Zhen, W.; Geng, Y.; Wang, Z.; Guo, Y. Pretreatment with probiotic Enterococcus faecium NCIMB 11181 ameliorates necrotic enteritis-induced intestinal barrier injury in broiler chickens. Sci. Rep. 2019, 9, 10256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovenský, J.; Švík, K.; Maťha, V.; Ištok, R.; Kamarád, V.; Ebringer, L.; Ferenčík, M.; Stančíková, M. Combination Treatment of Rat Adjuvant-Induced Arthritis with Methotrexate, Probiotic Bacteria Enterococcus faecium, and Selenium. Ann. N. Y. Acad. Sci. 2005, 1051, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Divyashri, G.; Krishna, G.; Muralidhara; Prapulla, S.G. Probiotic attributes, antioxidant, anti-inflammatory and neuromodulatory effects of Enterococcus faecium CFR 3003: In vitro and in vivo evidence. J. Med. Microbiol. 2015, 64, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Pitsouni, E.; Alexiou, V.; Saridakis, V.; Peppas, G.; Falagas, M.E. Does the use of probiotics/synbiotics prevent postoperative infections in patients undergoing abdominal surgery? A meta-analysis of randomized controlled trials. Eur. J. Clin. Pharmacol. 2009, 65, 561–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukacs, S.L.; Schoendorf, K.C.; Schuchat, A. Trends in Sepsis-Related Neonatal Mortality in the United States, 1985–1998. Pediatr. Infect. Dis. J. 2004, 23, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Wynn, J.L.; Scumpia, P.O.; Delano, M.J.; O’Malley, K.A.; Ungaro, R.; Abouhamze, A.; Moldawer, L.L. Increased mortality and altered immunity in neonatal sepsis produced by generalized peritonitis. Shock 2007, 28, 675–683. [Google Scholar] [CrossRef]
- Angurana, S.K.; Bansal, A.; Singhi, S.; Aggarwal, R.; Jayashree, M.; Salaria, M.; Mangat, N.K. Evaluation of Effect of Probiotics on Cytokine Levels in Critically Ill Children With Severe Sepsis: A Double-Blind, Placebo-Controlled Trial. Crit. Care Med. 2018, 46, 1656–1664. [Google Scholar] [CrossRef]
- Yılmaz, M.; Erdem, A.O. The Protective Role of Probiotics in Sepsis Induced Rats. Turk. J. Trauma Emerg. Surg. 2020, 26, 843–846. [Google Scholar] [CrossRef]
- Feng, G.; Dong, S.; Huang, M.; Zeng, M.; Liu, Z.; Zhao, Y.; Wu, H. Biogenic Polyphosphate Nanoparticles from a Marine Cyanobacterium Synechococcus sp. PCC 7002: Production, Characterization, and Anti-Inflammatory Properties In Vitro. Mar. Drugs 2018, 16, 322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasana, W.P.; Senevirathne, A.; Nikapitiya, C.; Eom, T.-Y.; Lee, Y.; Lee, J.-S.; Kang, D.-H.; Oh, C.; De Zoysa, M. A Novel Pseudoalteromonas xiamenensis Marine Isolate as a Potential Probiotic: Anti-Inflammatory and Innate Immune Modulatory Effects against Thermal and Pathogenic Stresses. Mar. Drugs 2021, 19, 707. [Google Scholar] [CrossRef]
- Hongpattarakere, T.; Cherntong, N.; Wichienchot, S.; Kolida, S.; Rastall, R.A. In vitro prebiotic evaluation of exopolysaccharides produced by marine isolated lactic acid bacteria. Carbohydr. Polym. 2012, 87, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Honkanen, P. Consumer Acceptance of (Marine) Functional Food; Academic Publishers: Wageningen, The Netherlands, 2009; pp. 141–154. [Google Scholar]
- Otaru, N.; Ye, K.; Mujezinovic, D.; Berchtold, L.; Constancias, F.; Cornejo, F.A.; Krzystek, A.; de Wouters, T.; Braegger, C.; Lacroix, C.; et al. GABA Production by Human Intestinal Bacteroides spp.: Prevalence, Regulation, and Role in Acid Stress Tolerance. Front. Microbiol. 2021, 12, 656895. [Google Scholar] [CrossRef] [PubMed]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef] [Green Version]
- De Vuyst, L.; Moreno, M.F.; Revets, H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 2003, 84, 299–318. [Google Scholar] [CrossRef]
- Allen, H.K.; Trachsel, J.; Looft, T.; Casey, T.A. Finding alternatives to antibiotics. Ann. N. Y. Acad. Sci. 2014, 1323, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Bs, S.; Thankappan, B.; Mahendran, R.; Muthusamy, G.; Selta, D.R.F.; Angayarkanni, J. Evaluation of GABA Production and Probiotic Activities of Enterococcus faecium BS5. Probiotics Antimicrob. Proteins 2021, 13, 993–1004. [Google Scholar] [CrossRef]
- Khanlari, Z.; Moayedi, A.; Ebrahimi, P.; Khomeiri, M.; Sadeghi, A. Enhancement of γ-aminobutyric acid (GABA) content in fermented milk by using Enterococcus faecium and Weissella confusa isolated from sourdough. J. Food Process. Preserv. 2021, 45, e15869. [Google Scholar] [CrossRef]
- Abriouel, H.; Ben Omar, N.; Molinos, A.C.; López, R.L.; Grande, M.J.; Martínez-Viedma, P.; Ortega, E.; Cañamero, M.M.; Galvez, A. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 2008, 123, 38–49. [Google Scholar] [CrossRef]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef]
- Zhou, N.; Zhang, J.; Fan, M.; Wang, J.; Guo, G.; Wei, X. Antibiotic resistance of lactic acid bacteria isolated from Chinese yogurts. J. Dairy Sci. 2012, 95, 4775–4783. [Google Scholar] [CrossRef]
- Pimentel, L.L.; Semedo, T.; Tenreiro, R.; Crespo, M.T.B.; Pintado, M.M.E.; Malcata, F.X. Assessment of Safety of Enterococci Isolated throughout Traditional Terrincho Cheesemaking: Virulence Factors and Antibiotic Susceptibility. J. Food Prot. 2007, 70, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- du Toit, M.; Franz, C.; Dicks, L.; Schillinger, U.; Haberer, P.; Warlies, B.; Ahrens, F.; Holzapfel, W. Characterisation and selection of probiotic lactobacilli for a preliminary minipig feeding trial and their effect on serum cholesterol levels, faeces pH and faeces moisture content. Int. J. Food Microbiol. 1998, 40, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Radulović, Z.; Petrović, T.; Nedović, V.; Dimitrijević, S.; Mirković, N.; Petrušić, M.; Paunović, D. Characterization of autochthonous Lactobacillus paracasei strains on potential probiotic ability. J. Dairy Prod. Process. Improv. 2010, 60, 86–93. [Google Scholar]
- Tannock, G.W. A Special Fondness for Lactobacilli. Appl. Environ. Microbiol. 2004, 70, 3189–3194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todorov, S.D.; Prévost, H.; Lebois, M.; Dousset, X.; LeBlanc, J.G.; Franco, B.D. Bacteriocinogenic Lactobacillus plantarum ST16Pa isolated from papaya (Carica papaya)—From isolation to application: Characterization of a bacteriocin. Food Res. Int. 2011, 44, 1351–1363. [Google Scholar] [CrossRef]
- Ding, L.; Gong, Y.; Yang, Z.; Zou, B.; Liu, X.; Zhang, B.; Li, J. Lactobacillus rhamnosus GG Ameliorates Liver Injury and Hypoxic Hepatitis in Rat Model of CLP-Induced Sepsis. Dig. Dis. Sci. 2019, 64, 2867–2877. [Google Scholar] [CrossRef]
- Panpetch, W.; Sawaswong, V.; Chanchaem, P.; Ondee, T.; Dang, C.P.; Payungporn, S.; Tumwasorn, S.; Leelahavanichkul, A. Candida Administration Worsens Cecal Ligation and Puncture-Induced Sepsis in Obese Mice Through Gut Dysbiosis Enhanced Systemic Inflammation, Impact of Pathogen-Associated Molecules From Gut Translocation and Saturated Fatty Acid. Front. Immunol. 2020, 11, 561652. [Google Scholar] [CrossRef]
- Ahmad, A.; Druzhyna, N.; Szabo, C. Delayed Treatment with Sodium Hydrosulfide Improves Regional Blood Flow and Alleviates Cecal Ligation and Puncture (CLP)-Induced Septic Shock. Shock 2016, 46, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Talarmin, H.; Derbré, F.; Lefeuvre-Orfila, L.; Léon, K.; Droguet, M.; Pennec, J.-P.; Giroux-Metgès, M.-A. The diaphragm is better protected from oxidative stress than hindlimb skeletal muscle during CLP-induced sepsis. Redox Rep. 2016, 22, 218–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.-L.; Liu, D.; Ma, C.-B. Review on the Angiotensin-I-Converting Enzyme (ACE) Inhibitor Peptides from Marine Proteins. Appl. Biochem. Biotechnol. 2012, 169, 738–749. [Google Scholar] [CrossRef]
- Chen, H.; Yan, X.; Zhu, P.; Lin, J. Antioxidant activity and hepatoprotective potential of agaro-oligosaccharides in vitro and in vivo. Nutr. J. 2006, 5, 31. [Google Scholar] [CrossRef]
- Higashimura, Y.; Naito, Y.; Takagi, T.; Mizushima, K.; Hirai, Y.; Harusato, A.; Ohnogi, H.; Yamaji, R.; Inui, H.; Nakano, Y.; et al. Oligosaccharides from agar inhibit murine intestinal inflammation through the induction of heme oxygenase-1 expression. J. Gastroenterol. 2012, 48, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Farag, A.M.; Hassan, S.W.; Beltagy, E.A.; El-Shenawy, M.A. Optimization of production of anti-tumor l-asparaginase by free and immobilized marine Aspergillus terreus. Egypt. J. Aquat. Res. 2015, 41, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Pandian, S.R.K.; Deepak, V.; Sivasubramaniam, S.D.; Nellaiah, H.; Sundar, K. Optimization and purification of anticancer enzyme L-glutaminase from Alcaligenes faecalis KLU102. Biologia 2014, 69, 1644–1651. [Google Scholar] [CrossRef]
- Indarmawan, T.; Mustopa, A.Z.; Budiarto, B.R.; Tarman, K. Antibacterial Activity of Extracellular Protease Isolated From an Algicolous Fungus Xylaria psidii KT30 Against Gram-Positive Bacteria. HAYATI J. Biosci. 2016, 23, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-Y.; Zhang, S.; Li, J.; Huang, N.; Sun, J.; Li, B.-H.; Yang, J.; Li, Z.-F. Baicalein Attenuates Severe Polymicrobial Sepsis via lleviating Immune Dysfunction of T Lymphocytes and Inflammation. Chin. J. Integr. Med. 2022, 28, 711–718. [Google Scholar] [CrossRef]
- Wang, W.; Ji, X.; Yuan, C.; Dai, F.; Zhu, J.; Sun, M. A Method for Molecular Analysis of Catalase Gene Diversity in Seawater. Indian J. Microbiol. 2013, 53, 477–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.-F.; Wang, Y.-F.; Hou, Y.-H.; Shi, Y.-L.; Han, H.; Miao, M.; Wu, Y.-Y.; Liu, Y.-P.; Yue, X.-N.; Li, Y.-J. Cloning, expression and biochemical characterization of recombinant superoxide dismutase from Antarctic psychrophilic bacterium Pseudoalteromonas sp. ANT506. J. Basic Microbiol. 2015, 56, 753–761. [Google Scholar] [CrossRef]
- Ji, M.; Barnwell, C.V.; Grunden, A.M. Characterization of recombinant glutathione reductase from the psychrophilic Antarctic bacterium Colwellia psychrerythraea. Extremophiles 2015, 19, 863–874. [Google Scholar] [CrossRef]
- Reboldi, A.; Dang, E.V.; Mcdonald, J.G.; Liang, G.; Russell, D.W.; Cyster, J.G. 25-Hydroxycholesterol suppresses interleukin-1–driven inflammation downstream of type I interferon. Science 2014, 345, 679–684. [Google Scholar] [CrossRef] [Green Version]
- Yin, K.; Hock, C.E.; Lai, P.-S.; Ross, J.T.; Yue, G. Role Of Interferon-Gamma In Lung Inflammation Following Cecal Ligation And Puncture In Rats. Shock 1999, 12, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-F.; Sun, C.-C.; Duan, J.-X.; Yang, H.-H.; Zhang, C.-Y.; Xiong, J.-B.; Zhong, W.-J.; Zu, C.; Guan, X.-X.; Jiang, H.-L.; et al. A COX-2/sEH dual inhibitor PTUPB ameliorates cecal ligation and puncture-induced sepsis in mice via anti-inflammation and anti-oxidative stress. Biomed. Pharmacother. 2020, 126, 109907. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.A.; Zhang, M.; Yin, F. Heme Oxygenase-1, Oxidation, Inflammation, and Atherosclerosis. Front. Pharmacol. 2012, 3, 119. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q. Role of Nrf2 in Oxidative Stress and Toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [Green Version]
- Qi, Z.; Wang, R.; Liao, R.; Xue, S.; Wang, Y. Neferine Ameliorates Sepsis-Induced Myocardial Dysfunction Through Anti-Apoptotic and Antioxidative Effects by Regulating the PI3K/AKT/mTOR Signaling Pathway. Front. Pharmacol. 2021, 12, 706251. [Google Scholar] [CrossRef]
- Sang, Z.; Dong, S.; Zhang, P.; Wei, Y. miR-214 ameliorates sepsis-induced acute kidney injury via PTEN/AKT/mTOR-regulated autophagy. Mol. Med. Rep. 2021, 24, 683. [Google Scholar] [CrossRef]
- Heemskerk, S.; Masereeuw, R.; Russel, F.G.M.; Pickkers, P. Selective iNOS inhibition for the treatment of sepsis-induced acute kidney injury. Nat. Rev. Nephrol. 2009, 5, 629–640. [Google Scholar] [CrossRef]
- Lei, W.; Liu, D.; Sun, M.; Lu, C.; Yang, W.; Wang, C.; Cheng, Y.; Zhang, M.; Shen, M.; Yang, Z.; et al. Targeting STAT3: A crucial modulator of sepsis. J. Cell. Physiol. 2021, 236, 7814–7831. [Google Scholar] [CrossRef]
- Zaghloul, E.H.; Ibrahim, H.A.H.; El-Badan, D.E.S. Production of biocement with marine bacteria; Staphylococcus epidermidis EDH to enhance clay water retention capacity. Egypt. J. Aquat. Res. 2020, 47, 53–59. [Google Scholar] [CrossRef]
- Amer, M.S.; Zaghloul, E.H.; Ibrahim, M.I.A. Characterization of exopolysaccharide produced from marine-derived Aspergillus terreus SEI with prominent biological activities. Egypt. J. Aquat. Res. 2020, 46, 363–369. [Google Scholar] [CrossRef]
- Guttmann, D.M.; Ellar, D.J. Phenotypic and genotypic comparisons of 23 strains from the Bacillus cereus complex for a selection of known and putative B. thuringiensis virulence factors. FEMS Microbiol. Lett. 2000, 188, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-Wide Assessment of Antibiotic Resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, N.; Juan, W.; Aiping, Z.; ChaoFeng, M.; Xiaokang, W.; Jiru, X.; Nawaz, M.; Wang, J.; Zhou, A.; Wu, X.; et al. Screening and characterization of new potentially probiotic lactobacilli from breast-fed healthy babies in Pakistan. Afr. J. Microbiol. Res. 2011, 5, 1428–1436. [Google Scholar] [CrossRef]
- Harsa, H.Ş.; Yavuzdurmaz, H. Isolation, Characterization, Determination of Probiotic Properties of Lactic Acid Bacteria from Human Milk. MSc. Thesis, Izmir Institute of Technology, Urla, Turkey, October 2007. [Google Scholar]
- Zubrow, M.E.; Margulies, S.S.; Yehya, N. Nordihydroguaiaretic acid reduces secondary organ injury in septic rats after cecal ligation and puncture. PLoS ONE 2020, 15, e0237613. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]







| Test | Result | Test | Result |
|---|---|---|---|
| Alpha-mannosidase (AMAN) | − | d-mannosE (dMNE) | + |
| Phosphatase (PHOS) | − | Methyl-b-d-glucopyranoside (MBdG) | + |
| Leucine arylamidase (LeuA) | − | Pullulan (PUL) | − |
| l-proline arylamidase (ProA) | − | d-raffinose (dRAF) | − |
| Tyrosine arylamidase (TyrA) | − | Optochin resistance (OPTO) | + |
| d-sorbitol (dSOR) | − | Urease (URE) | − |
| Polymixin b resistance (POLYB) | + | d-mannitol (dMAN) | − |
| d-amygdalin (AMY) | − | d-galactose (dGAL) | + |
| Phosphatidylinositol phospholipase (PIPLC) | − | d-ribose (dRIB) | + |
| d-xylose (dXYL) | − | l-lactate alkalinization (ILATK) | − |
| Arginine dihydrolase 1 (ADH1) | + | Lactose (LAC) | + |
| Beta-galactosidase (BGAL) | + | N-acetyl-d-glucosamine (NAG) | + |
| Alpha-glucosidase (AGLU) | − | d-maltose (dMAL) | + |
| Beta glucuronidase (BGURr) | − | O/129 resistance (comp. vibrio.) (O129R) | + |
| Alpha-galactosidase (AGAL) | − | Salicin (SAL) | + |
| l-pyrrolydonyl-arylamidase (PyrA) | + | Saccharose/Sucrose (SAC) | − |
| Beta-glucuronidase (BGUR) | − | d-trehalose (dTRE) | + |
| Alanine arylamidase (AlaA) | − | Arginine dihydrolase 2 (ADH2s) | + |
| Ala-phe-pro arylamidase (APPA) | − | Bacitracin resistance (BACI) | + |
| Cyclodextrin (CDEX) | + | Novobiocin resistance (NOVO) | + |
| l-aspartate arylamidase (AspA) | − | Growth in 6.5% NaCl (NC6.5) | + |
| Beta galactopyranosidase (BGAR) | − |
| Antibiotic | Result |
|---|---|
| Nalidixic Acid (30 mcg) | R |
| Ampicillin (10 mcg) | R |
| Erythromycin (15 mcg) | R |
| Tetracycline (30 mcg) | S |
| Piperacillin/Tazobactam (110 mcg) | R |
| Vancomycin (30 mcg) | S |
| Oxacillin (1 mcg) | S |
| Ceftriaxone (30 mcg) | R |
| Amoxicillin (25 mcg) | S |
| Ofloxacin (5 mcg) | S |
| Cephradine (5 mcg) | S |
| Indicator Pathogen | Inhibition Zone Diameter (mm) |
|---|---|
| Pseudomonas fluorescens ATCC 13525 | 18 ± 0.5 |
| Streptococcus agalactiae ATCC 13813 | 19 ± 0.44 |
| Aeromonas hydrophila ATCC 13037 | 18 ± 0.23 |
| Staphylococcus aureus ATCC 25923 | 21 ± 0.115 |
| Escherichia coli ATCC 8739 | 15 ± 0.05 |
| Enterococcus faecalis ATCC 29212 | 18 ± 0.15 |
| Klebsiella pneumonia ATCC 13883 | 15 ± 0.23 |
| SHAM | SHAM + EA9 | CLP | CLP + EA9 | |
|---|---|---|---|---|
| AST | 143.83 ± 13.79 ** | 141.16 ± 20.06 ** | 211 ± 25.49 | 164.5 ± 41.92 * |
| ALT | 57.33 ± 8.94 ** | 55.33 ± 7.89 ** | 83.5 ± 13.14 | 63 ± 12.17 * |
| BUN | 72.16 ± 14.77 * | 72.51 ± 16.00 * | 104 ± 28.09 | 74.66 ± 7.89 * |
| Creatinine | 0.71 ± 0.09 * | 0.71 ± 0.08 * | 1.1 ± 0.33 | 0.73 ± 0.15 * |
| Gene Name | Accession Number | Sequences |
|---|---|---|
| Interleukin-10 (IL-10) | XM_006249712.4 | F: 5′-TGCCTTCAGTCAAGTGAAGAC-3′ |
| R: 5′-AAACTCATTCATGGCCTTGTA-3′ | ||
| Interleukin-1β (IL-1β) | NM_031512.2 | F: 5′-CACCTCTCAAGCAGAGCACAG-3′ |
| R: 5′-GGGTTCCATGGTGAAGTCAAC-3′ | ||
| Interferon-γ (INF-γ) | NM_138880.3 | F: 5′-GTGTCATCGAATCGCACCTG-3′ |
| R: 5′-GTTCACCTCGAACTTGGCGA-3′ | ||
| Cyclooxygenase-2 (COX-2) | S67722.1 | F: 5′-TGAGTACCGCAAACGCTTCT-3′ |
| R: 5′-ACACAGGAATCTTCACAAATGGA-3′ | ||
| Superoxide dismutase-1 (SOD-1) | NM_017050.1.1 | F: 5′-TAACTGAAGGCGAGCATGGG-3′ |
| R: 5′-CCTCTCTTCATCCGCTGGAC-3′ | ||
| Superoxide dismutase-2 (SOD-2) | NM_017051.2 | F: 5′-AATCAACAGACCCAAGCTAGGC-3′ |
| R: 5′-CACAATGTCACTCCTCTCCGAA-3′ | ||
| Heme oxygenase-1 (HO-1) | XM_039097470.1 | F: 5′-GTAAATGCAGTGTTGGCCCC-3′ |
| R: 5′-ATGTGCCAGGCATCTCCTTC-3′ | ||
| Nuclear factor erythroid 2-related factor-2 (Nrf-2) | NM_031789.3 | F: 5′-TTGTAGATGACCATGAGTCGC-3′ |
| R: 5′-TGTCCTGCTGTATGCTGCTT-3′ | ||
| Protein kinase B (AKT) | XM_006240631.3 | F: 5′-ACCTCTGAGACCGACACCAG-3′ |
| R: 5′-AGGAGAACTGGGGAAAGTGC-3′ | ||
| Mammalian target of rapamycin (mTOR) | NM_019906.2 | F: 5′-GACAACAGCCAGGGCCGCAT-3′ |
| R: 5′-ACGCTGCCTTTCTCGACGGC-3′ | ||
| Inducible nitric oxide synthase (iNOS) | XM_039085203.1 | F: 5′-ATGGAACAGTATAAGGCAAACACC-3′ |
| R: 5′-GTTTCTGGTCGATGTCATGAGCAAAGG-3′ | ||
| Signal transducer and activator of transcription-3 (STAT-3) | XM_006247257.4 | F: 5′-GGGCCTGGTGTGAACTACTC-3′ |
| R: 5′-ATGGTATTGCTGCAGGTCGT-3′ | ||
| Beta actin (β-actin) | NM_031144.3 | F: 5′-CCGCGAGTACAACCTTCTTG-3′ |
| R: 5′-CAGTTGGTGACAATGCCGTG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaghloul, E.H.; Abuohashish, H.M.; El Sharkawy, A.S.; Abbas, E.M.; Ahmed, M.M.; Al-Rejaie, S.S. Probiotic Potential of the Marine Isolate Enterococcus faecium EA9 and In Vivo Evaluation of Its Antisepsis Action in Rats. Mar. Drugs 2023, 21, 45. https://doi.org/10.3390/md21010045
Zaghloul EH, Abuohashish HM, El Sharkawy AS, Abbas EM, Ahmed MM, Al-Rejaie SS. Probiotic Potential of the Marine Isolate Enterococcus faecium EA9 and In Vivo Evaluation of Its Antisepsis Action in Rats. Marine Drugs. 2023; 21(1):45. https://doi.org/10.3390/md21010045
Chicago/Turabian StyleZaghloul, Eman H., Hatem M. Abuohashish, Amany S. El Sharkawy, Eman M. Abbas, Mohammed M. Ahmed, and Salim S. Al-Rejaie. 2023. "Probiotic Potential of the Marine Isolate Enterococcus faecium EA9 and In Vivo Evaluation of Its Antisepsis Action in Rats" Marine Drugs 21, no. 1: 45. https://doi.org/10.3390/md21010045
APA StyleZaghloul, E. H., Abuohashish, H. M., El Sharkawy, A. S., Abbas, E. M., Ahmed, M. M., & Al-Rejaie, S. S. (2023). Probiotic Potential of the Marine Isolate Enterococcus faecium EA9 and In Vivo Evaluation of Its Antisepsis Action in Rats. Marine Drugs, 21(1), 45. https://doi.org/10.3390/md21010045