Pentaketides and 5-p-Hydroxyphenyl-2-pyridone Derivative from the Culture Extract of a Marine Sponge-Associated Fungus Hamigera avellanea KUFA0732
Abstract
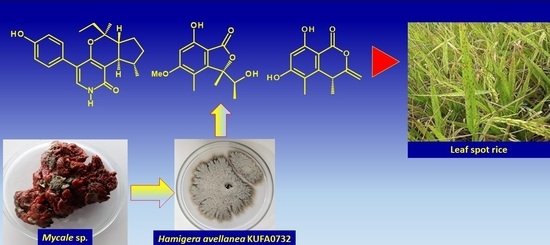
:1. Introduction
2. Results and Discussions
3. Methods and Materials
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Extraction and Isolation
3.3.1. (R)-6,8-Dihydroxy-4,5-dimethyl-3-methylidene-3,4-dihydro-1H-2-benzopyran-1-one (1)
3.3.2. (3S,4R)-3,8-Dihydroxy-6-methoxy-4,5-dimethyl-1-oxo-3,4-dihydro-1H-2-benzopyran-3-yl)methyl acetate (2)
3.3.3. (R)-5, 7-Dimethoxy-3-((S)-(1-hydroxyethyl)-3,4-dimethylisobenzofuran 1(3H)-one (4b)
3.3.4. (S)-7-Hydroxy-3-((S)-1-hydroxyethyl)-5-methoxy-3,4-dimethylisobenzofuran 1(3H)-one (5)
3.3.5. Avellaneanone (6)
3.4. X-ray Crystal Structures
3.4.1. X-ray Crystal Structure of 1
3.4.2. X-ray Crystal Structure of 4b
3.4.3. X-ray Crystal Structure of 5
3.4.4. X-ray Crystal Structure of 6
3.5. Antifungal Activity Bioassays
3.5.1. Antifungal Activity of the Crude Extract against Plant Pathogenic Fungi
3.5.2. Antifungal Activity of Isolated Compounds against Plant Pathogenic Fungi
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peterson, S.W.; Jurjevic, Z.; Bills, G.F.; Stchigel, A.M.; Guarro, J.; Vega, F.E. Genus Hamigera, six new species and multilocus DNA sequence based phylogeny. Mycologia 2010, 102, 847–864. [Google Scholar] [CrossRef]
- Igarashia, Y.; Hanafusa, T.; Gohda, F.; Peterson, S.; Bills, G. Species-level assessment of secondary metabolite diversity among Hamigera species and a taxonomic note on the genus. Mycology 2014, 5, 102–109. [Google Scholar] [CrossRef]
- Yamazaki, M.; Horie, Y.; Bae, K.; Maebayashi, Y.; Jisai, Y.; Fujimoto, H. New fungal metabolites avellanins A and B from Hamigera avellanea, with pressure effect. Chem. Pharm. Bull. 1987, 35, 2122–2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breinholt, J.; Kjaer, A.; Olsen, C.E.; Rassing, B.R. A bis-formamidodiphenylbutadiene from the fungus Hamigera avellanea. Acta Chem. Scand. 1996, 50, 643–645. [Google Scholar] [CrossRef]
- Breinholt, J.; Kjaer, A.; Olsen, C.E.; Rassing, B.R.; Rosendahl, C.N. Hamigerone and dihydrohamigerone: Two acetate-derived, antifungal metabolites from Hamigera avellanea. Acta Chem. Scand. 1997, 51, 1241–1244. [Google Scholar] [CrossRef] [Green Version]
- Isaka, M.; Chinthanom, P.; Veeranondha, S.; Supothina, S.; Luangsa-ard, J.J. Novel cyclopropyl diketones and 14-membered macrol ides from the soil fungus Hamigera avellanea BCC 17816. Tetrahedron 2008, 64, 11028–11033. [Google Scholar] [CrossRef]
- Isaka, M.; Chinthanom, P.; Kongthong, S.; Supothina, S.; Ittiworapong, S. Hamigeromycins C–G, 14-membered macrolides from the fungus Hamigera avellanea BCC 17816. Tetrahedron 2010, 66, 955–961. [Google Scholar] [CrossRef]
- Chalearmsrimuang, T.; Ismail, S.I.; Mazlan, N.; Suasaard, S.; Dethoup, T. Marine-derived fungi: A promising source of halotolerant biological control agents against plant pathogenic fungi. J. Pure Appl. Microbiol. 2019, 13, 209–223. [Google Scholar] [CrossRef] [Green Version]
- Dethoup, T.; Kaewsalong, N.; Songkumorn, P.; Jantasorn, A. Potential application of a marine-derived fungus, Talaromyces tratensis KUFA 0091 against rice diseases. Biol. Control 2018, 119, 1–6. [Google Scholar] [CrossRef]
- Klaram, R.; Jantasorn, A.; Dethoup, T. Efficacy of marine antagonist, Trichoderma spp. as halo-tolerant biofungicide in controlling rice diseases and yield improvement. Biol. Control 2022, 172, 104985. [Google Scholar] [CrossRef]
- Chinworrungsee, M.; Kittakoop, P.; Isaka, M.; Chanphen, R.; Tanticharoen, M.; Thebtaranonth, Y. Halorosellins A and B, unique isocoumarin glucosides from the marine fungus Halorosellinia oceanica. J. Chem. Soc. Perkin Trans. 1 2002, 2473–2476. [Google Scholar] [CrossRef]
- Bi, Y.M.; Bi, X.B.; Fang, A.; Zhao, Q.R. Metabolites from the fungus Cephalosporium sp. AL031. Arch. Pharm. Res. 2007, 30, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Tayone, W.C.; Honma, M.; Kanamaru, S.; Noguchi, S.; Tanaka, K.; Nehira, T.; Hashimoto, M. Stereochemical investigations of isochromenones and isobenzofuranones isolated from Leptosphaeria sp. KTC 727. J. Nat. Prod. 2011, 74, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Aoki, R. Isosclerone, a new metabolite of Sclerotinia sclerotiorum (LIB.) DE BARY. Agric. Biol. Chem. 1974, 38, 1501–1505. [Google Scholar] [CrossRef]
- Evidente, A.; Sparapano, L.; Anna AndolfiI, A.; Bruno, G. Two naphthalenone pentaketides from liquid cultures of Phaeoacremonium aleophilum, a fungus associated with esca of grapevine. Phytopathol. Mediterr. 2000, 39, 162–168. [Google Scholar]
- Husain, S.M.; Müller, M. Fungal dihydroxynaphthalene-melanin: Diversity-Oriented biosynthesis through enzymatic and non-enzymatic transformations. Synlett 2017, 28, 2360–2372. [Google Scholar] [CrossRef]
- Tayone, W.C.; Kanamaru, S.; Honma, M.; Tanaka, K.; Nehira, T.; Hashimoto, M. Absolute stereochemistry of novel isochromanone derivatives from Leptosphaeria sp. KTC 727. Biosci. Biotechnol Biochem. 2011, 75, 2390–2393. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.X.; Wang, J.; Li, S.W.; Cheng, B.; Li, L.; Chen, B.; Liu, L.; Lin, Y.C.; Gu, Y.C. Metabolites of the mangrove fungus Xylaria sp. BL321 from the South China Sea. Planta Med. 2012, 78, 172–176. [Google Scholar] [CrossRef] [Green Version]
- Wat, C.-K.; Mcinnes, A.G.; Smith, D.G.; Wright, J.L.C.; Vining, L.C. The yellow pigments of Beauveria species. Structures of tenellin and bassianin. Can. J. Chem. 1977, 55, 4090–4098. [Google Scholar] [CrossRef] [Green Version]
- Heneghan, M.N.; Yakasai, A.A.; Williams, K.; Kadir, K.A.; Wasil, Z.; Bakeer, W.; Fisch, K.M.; Bailey, A.M.; Simpson, T.J.; Cox, R.J.; et al. The programming role of trans-acting enoyl reductases during the biosynthesis of highly reduced fungal polyketides. Chem. Sci. 2011, 2, 972–979. [Google Scholar] [CrossRef]
- Bergmann, S.; Schümann, J.; Scherlach, K.; Lange, C.; Brakhage, A.A.; Hertweck, C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus Nidulans. Nat. Chem. Biol. 2007, 3, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-C.; Lee, Y.-W.; Tamura, H.; Yoshizawa, T. Sambutoxin: A new mycotoxin isolated from Fusarium sambucinum. Tetrahedron Lett. 1995, 36, 1047–1050. [Google Scholar] [CrossRef]
- Matsumoto, M.; Minato, H. Structure of ilicicolin H, an antifungal antibiotic. Tetrahedron Lett. 1976, 42, 3827–3830. [Google Scholar] [CrossRef]
- Wu, B.; Oesker, V.; Wiese, J.; Schmaljohann, R.; Imhoff, J.F. Two new antibiotic pyridones produced by a marine fungus, Trichoderma sp. Strain MF106. Mar. Drugs 2014, 12, 1208–1219. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Contesini, F.J.; Wang, X.; Ghidinelli, S.; Tornby, D.S.; Andersen, T.E.; Mortensen, U.F.; Larsen, T.O. Biosynthesis of calipyridone A represents a fungal 2-pyridone formation without ring expansion in Aspergillus californicus. Org. Lett. 2022, 24, 804–808. [Google Scholar] [CrossRef]
- Fisch, K.M.; Bakeer, W.; Yakasai, A.A.; Song, Z.; Pedrick, J.; Wasil, Z.; Bailey, A.M.; Lazarus, C.M.; Simpson, T.J.; Cox, R.J. Rational domain swaps decipher programming in fungal highly reducing polyketide synthases and resurrect an extinct metabolite. J. Am. Chem. Soc. 2011, 133, 16635–16641. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 72, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, A64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline; CLSI Document M26-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999. [Google Scholar]











| Position | δC, Type | δH, (J in Hz) | COSY | HMBC |
|---|---|---|---|---|
| 1 | 166.3, C | |||
| 3 | 157.4, C | |||
| 4 | 34.2, CH | 4.02 q (7.2) | H3-10 | C-4a, 5, 8a, 11 |
| 4a | 143.6, C | |||
| 5 | 113.2, C | |||
| 6 | 164.1, C | |||
| 7 | 101.0, CH | 6.33,s | H3-11 | C-5, 6, 8, 8a |
| 8 | 161.8, C | |||
| 8a | 97.7, C | |||
| 9 | 96.2, CH2 | 4.75, d (1.6) 4.73, d (1.6) | C-3, 4 | |
| 10 | 10.2, CH3 | 1.27, d (7.0) | H-4 | C-3, 4, 4a |
| 11 | 22.3 CH3 | 2.02, s | H-7 | C-4a, 5, 6 |
| OH-8 | - | 10.70, brs |
| Position | δC, Type | δH, (J in Hz) | COSY | HMBC | ROESY |
|---|---|---|---|---|---|
| 1 | 168.2, CO | ||||
| 3 | 102.2, C | ||||
| 4 | 35.9, CH | 4.33, q (7.1) | H3-10, | C-3, 4a, 5, 8a, 10 | H3-10, H-11 |
| 4a | 141.5, C | ||||
| 5 | 115.5, C | ||||
| 6 | 164.9, C | ||||
| 7 | 97.5, CH | 6.37, s | C-3, 5, 8, 8a, 12 | OMe-6 | |
| 8 | 163.1, C | ||||
| 8a | 99.0, C | ||||
| 9a b | 65.9, CH2 | 4.25, d (11.8) 4.59, d (11.8) | H-9b H-9a | C-12 C-3, 12 | H3-10 H3-10 |
| 10 | 16.2, CH3 | 1.19, d (7.1) | H-4 | C-3, 4, 4a | H-9a, 9b |
| 11 | 10.1, CH3 | 2.08, s | C-3, 4a, 6 | H-4 | |
| 12 | 170.7, CO | ||||
| 13 | 20.8, CH3 | 2.19, s | C-12 | ||
| OMe-6 | 55.9, CH3 | 3.85, s | C-6 | H-7 | |
| OH-8 | - | 11.14, s | C-7, 8 |
| Position | δC, Type | δH, (J in Hz) | COSY | HMBC |
|---|---|---|---|---|
| 1 | 168.1, CO | |||
| 3 | 88.7, C | |||
| 3a | 152.8, C | |||
| 4 | 111.7, C | |||
| 5 | 164.5, C | |||
| 6 | 94.5, CH | 6.04, s | OMe-5, H3-11 | C-1, 4, 5, 7, 7a |
| 7 | 158.3, C | |||
| 7a | 105.4, C | |||
| 8 | 70.9, CH | 4.20, m | H3-9 | |
| 9 | 17.8, CH3 | 0.86, d (6.5) | H-8 | C-3, 8 |
| 10 | 21.5, CH3 | 1.74, s | C-3, 3a, 8 | |
| 11 | 11.2, CH3 | 2.10, s | C-3a, 4, 5 | |
| OMe-5 | 56.0, CH3 | 3.91, s | C-5 | |
| OMe-7 | 56.1, CH3 | 3.96, s | C-7 |
| Position | δC, Type | δH, (J in Hz) | COSY | HMBC |
|---|---|---|---|---|
| 1 | 171.1, CO | |||
| 3 | 91.6, C | |||
| 3a | 149.8, C | |||
| 4 | 112.4, C | |||
| 5 | 165.3, C | |||
| 6 | 98.2, CH | 6.43, s | H3-11, OMe-5 | C-1, 4,7, 7a |
| 7 | 156.3, C | |||
| 7a | 103.7, C | |||
| 8 | 70.5, CH | 4.26, m | H3-9 | |
| 9 | 17.9, CH3 | 1.40, d (6.4) | C-3, 8 | |
| 10 | 21.1, CH3 | 1.67, s | C-3, 3a, 8 | |
| 11 | 11.1, CH3 | 2.17,s | H-6 | C-3a, 4, 5 |
| OMe-5 | 56.3, CH3 | 3.87, s | H-6 | C-5 |
| OH-7 | 7.67, brs |
| Position | δC, Type | δH, (J in Hz) | COSY | HMBC |
|---|---|---|---|---|
| NH-1 | - | 10.93, brs | ||
| 2 | 162.4 CO | |||
| 3 | 110.4 C | |||
| 4 | 160.2, C | |||
| 5 | 113.6, C | |||
| 6 | 131.5, CH | 6.98,s | C-2, 4.5, 17 | |
| 7 | 43.8, CH | 2.06, dd (12.0, 8.7) | H-8, 11 | C-3, 11, 12 |
| 8 | 51.9, CH | 1.71, ddd (12.0, 12.0, 7.0) | H-7, 9 | C-13, 14 |
| 9 | 25.2 CH2 | 1.56, m | H-8, 10 | |
| 10 | 34.5, CH2 | 1.39, m 1.99, m | H-9, 11 | C-9, 11 |
| 11 | 35.3, CH | 2.15, m | H3-12 | |
| 12 | 25.1, CH3 | 1.47, d (6.2) | H-11 | C-7, 10, 11 |
| 13 | 82.2, C | - | ||
| 14 | 18.6, CH3 | 1.11, s | C-8, 13, 15 | |
| 15 | 33.8, CH2 | 1.55, q (7.1) | H3-16 | |
| 16 | 7.7, CH3 | 0.85, t (7.3) | H-15 | C-13, 15 |
| 17 | 126.7, C | - | ||
| 18 | 130.6, CH | 7.15, d (8.6) | H-19 | H-5, 20, 22 |
| 19 | 115.1, CH | 6.73, d (8.6) | H-18 | H-17, 20, 21 |
| 20 | 156.6, C | - | ||
| 21 | 115.1, CH | 6.73, d (8.6) | H-22 | H-17, 19, 20 |
| 22 | 130.6, CH | 7.15, d (8.6) | H-21 | H-5, 18, 20 |
| 23 | OH-20 | 9.38, brs |
| Plant Pathogen | % Mycelial Growth Inhibition at Concentrations | |
|---|---|---|
| 10 g/L | 1 g/L | |
| Alternaria brassicicola (black spot of Chinese Kale) | 68.67 ± 2.27 c | 47.33 ± 1.62 d |
| Bipolaris oryzae (brown spot of rice) | 65.21 ± 3.12 d | 36.09 ± 1.14 f |
| Colletotrichum capsici (anthracnose of chili) | 100 ± 0 a | 58.67 ± 2.08 c |
| C. gloeosporiodes (anthracnose of mango) | 100 ± 0 a | 62.32 ± 3.58 b |
| Curvularia oryzae (leaf spot of rice) | 100 ± 0 a | 63.67 ± 2.15 b |
| Fusarium semitectum (dirty panicle of rice) | 85.34 ± 3.61 b | 38.04 ± 1.36 e |
| Lasiodiplodia theobromae (fruit rot of mangosteen) | 100 ± 0 a | 37.42 ± 1.08 e f |
| Phytophthora palmivora (root and stem rot of durian) | 100 ± 0 a | 0 ± 0 h |
| Pyricularia oryzae (rice blast) | 100 ± 0 a | 46.25 ± 2.01 d |
| Rhizoctonia oryzae (sheath blight of rice) | 100 ± 0 a | 68.34 ± 3.20 a |
| Sclerotium rolfsii (stem rot of bean) | 100 ± 0 a | 33.47 ± 1.06 g |
| Plant Pathogen | Plant Disease | MIC (µg/Ml) | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 4b | 5 | 6 | 7 | Mancozeb | ||
| Alternaria brassicicola | black spot of Chinese Kale | >500 | >500 | >500 | >500 | >500 | >500 | 125 |
| Bipolaris oryzae | brown spot of rice | >500 | >500 | >500 | >500 | >500 | >500 | 125 |
| Colletotrichum capsici | anthracnose of chili | >500 | >500 | >500 | >500 | >500 | >500 | 125 |
| C. gloeosporiodes | anthracnose of mango | >500 | 500 | >500 | >500 | >500 | >500 | 125 |
| Curvularia oryzae | leaf spot of rice | >500 | 125 | >500 | 125 | 250 | >500 | 125 |
| Fusarium semitectum | dirty panicle of rice | >500 | >500 | >500 | >500 | >500 | >500 | 125 |
| Lasiodiplodia theobromae | fruit rot of mangosteen | >500 | >500 | >500 | >500 | >500 | >500 | 250 |
| Phytophthora palmivora | root and stem of durian | >500 | >500 | >500 | >500 | >500 | >500 | 125 |
| Pyricularia oryzae | rice blast | >500 | >500 | >500 | >500 | >500 | >500 | 125 |
| Rhizoctonia oryzae | sheath blight of rice | >500 | >500 | >500 | >500 | >500 | >500 | 250 |
| Sclerotium rolfsii | stem rot of bean | >500 | >500 | >500 | >500 | >500 | >500 | 250 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klaram, R.; Dethoup, T.; Machado, F.P.; Gales, L.; Kumla, D.; Hafez Ghoran, S.; Sousa, E.; Mistry, S.; Silva, A.M.S.; Kijjoa, A. Pentaketides and 5-p-Hydroxyphenyl-2-pyridone Derivative from the Culture Extract of a Marine Sponge-Associated Fungus Hamigera avellanea KUFA0732. Mar. Drugs 2023, 21, 344. https://doi.org/10.3390/md21060344
Klaram R, Dethoup T, Machado FP, Gales L, Kumla D, Hafez Ghoran S, Sousa E, Mistry S, Silva AMS, Kijjoa A. Pentaketides and 5-p-Hydroxyphenyl-2-pyridone Derivative from the Culture Extract of a Marine Sponge-Associated Fungus Hamigera avellanea KUFA0732. Marine Drugs. 2023; 21(6):344. https://doi.org/10.3390/md21060344
Chicago/Turabian StyleKlaram, Rotchana, Tida Dethoup, Fátima P. Machado, Luís Gales, Decha Kumla, Salar Hafez Ghoran, Emília Sousa, Sharad Mistry, Artur M. S. Silva, and Anake Kijjoa. 2023. "Pentaketides and 5-p-Hydroxyphenyl-2-pyridone Derivative from the Culture Extract of a Marine Sponge-Associated Fungus Hamigera avellanea KUFA0732" Marine Drugs 21, no. 6: 344. https://doi.org/10.3390/md21060344
APA StyleKlaram, R., Dethoup, T., Machado, F. P., Gales, L., Kumla, D., Hafez Ghoran, S., Sousa, E., Mistry, S., Silva, A. M. S., & Kijjoa, A. (2023). Pentaketides and 5-p-Hydroxyphenyl-2-pyridone Derivative from the Culture Extract of a Marine Sponge-Associated Fungus Hamigera avellanea KUFA0732. Marine Drugs, 21(6), 344. https://doi.org/10.3390/md21060344