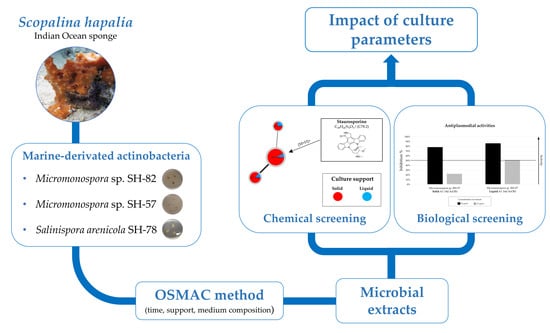
OSMAC Method to Assess Impact of Culture Parameters on Metabolomic Diversity and Biological Activity of Marine-Derived Actinobacteria
Abstract
:1. Introduction
2. Results
2.1. Micromonospora sp. SH-82
2.1.1. Influence of Culture Support and Time on Micromonospora sp. SH-82’s Productions
Chemical Analysis
Bioassays
2.1.2. Influence of Medium Composition and Culture Support on Micromonospora sp. SH-82’s Productions
Chemical Analysis
Bioassays
2.1.3. Overview of the Annotations
2.2. Micromonospora sp. SH-57
2.2.1. Influence of Culture Support and Time on Micromonospora sp. SH-57’s Productions
Chemical Analysis
Bioassays
2.2.2. Influence of Medium Composition and Culture Support on Micromonospora sp. SH-57’s Productions
2.2.3. Overview of the Annotations
2.3. Salinispora arenicola SH-78
2.3.1. Influence of Culture Support on Salinispora arenicola SH-78’s Productions
2.3.2. Influence of Culture Time on Salinispora arenicola SH-78’s Productions
2.3.3. Influence of Extraction Solvent on Salinispora arenicola SH-78’s Productions
Molecular Network
Bioassays
2.3.4. Overview of the Annotations
3. Discussion
3.1. Micromonospora sp. SH-82
- An extended cultivation time enhances the production of metabolites.
- A robust production, whatever the culture support.
- A1 medium boosts chemical diversity, while MB medium sustains bioactivity.
- Annotation of bioactive metabolites.
3.2. Micromonospora sp. SH-57
- The liquid support culture is essential for the production of metabolites.
- An extended cultivation time and A1 medium are still favorable.
- Annotation of bioactive metabolites.
3.3. Salinispora arenicola SH-78
- A variable influence of time and culture support on the production of metabolites.
- The use of different solvents increases the quantity and diversity of metabolites.
- Annotation of bioactive metabolites.
3.4. The Common Impact of Culture Parameters on the Production of Metabolites
4. Materials and Methods
4.1. Biological Material (Sponge/Microbial Strains)
4.2. Cultivation of Strains Using the “One Strain Many Compounds” (OSMAC) Method
4.3. Microbial Extract Preparation
4.4. Chemical Analysis
4.4.1. HPLC-DAD-CAD Analysis
4.4.2. UHPLC-QTOF-MS/MS Analysis
4.5. Raw Data Processing, Ion Identity Molecular Networks, and Annotations
4.6. Biological Activity Tests
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.-Y.; Li, H.-J.; Li, Q.-Y.; Wu, Y.-C. Application of Marine Natural Products in Drug Research. Bioorg. Med. Chem. 2021, 35, 116058. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Maharshi, A.; Jha, B. Actinobacteria in Natural Products Research: Progress and Prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds. Mar. Drugs 2021, 19, 530. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.S.U.; Shaikh, A.L. Marine Actinobacteria as a Drug Treasure House. Biomed. Pharmacother. 2017, 87, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, Y.; Bouk, S.; Kakaei, E.; Taheri, M. Natural Products from Actinobacteria as a Potential Source of New Therapies Against Colorectal Cancer: A Review. Front. Pharmacol. 2022, 13, 929161. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial Communication Leading to the Activation of Silent Fungal Secondary Metabolite Gene Clusters. Front. Microbiol. 2015, 6, 299. [Google Scholar] [CrossRef]
- Okada, B.K.; Seyedsayamdost, M.R. Antibiotic Dialogues: Induction of Silent Biosynthetic Gene Clusters by Exogenous Small Molecules. FEMS Microbiol. Rev. 2017, 41, 19–33. [Google Scholar] [CrossRef]
- Pinedo-Rivilla, C.; Aleu, J.; Durán-Patrón, R. Cryptic Metabolites from Marine-Derived Microorganisms Using OSMAC and Epigenetic Approaches. Mar. Drugs 2022, 20, 84. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Palma Esposito, F.; Giugliano, R.; Della Sala, G.; Vitale, G.A.; Buonocore, C.; Ausuri, J.; Galasso, C.; Coppola, D.; Franci, G.; Galdiero, M.; et al. Combining OSMAC Approach and Untargeted Metabolomics for the Identification of New Glycolipids with Potent Antiviral Activity Produced by a Marine Rhodococcus. Int. J. Mol. Sci. 2021, 22, 9055. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Influence of OSMAC-Based Cultivation in Metabolome and Anticancer Activity of Fungi Associated with the Brown Alga Fucus Vesiculosus. Mar. Drugs 2019, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Said Hassane, C.; Fouillaud, M.; Le Goff, G.; Sklirou, A.D.; Boyer, J.B.; Trougakos, I.P.; Jerabek, M.; Bignon, J.; de Voogd, N.J.; Ouazzani, J.; et al. Microorganisms Associated with the Marine Sponge Scopalina Hapalia: A Reservoir of Bioactive Molecules to Slow Down the Aging Process. Microorganisms 2020, 8, 1262. [Google Scholar] [CrossRef]
- Yan, S.; Zeng, M.; Wang, H.; Zhang, H. Micromonospora: A Prolific Source of Bioactive Secondary Metabolites with Therapeutic Potential. J. Med. Chem. 2022, 65, 8735–8771. [Google Scholar] [CrossRef] [PubMed]
- Haque, N.; Parveen, S.; Tang, T.; Wei, J.; Huang, Z. Marine Natural Products in Clinical Use. Mar. Drugs 2022, 20, 528. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The Marine Actinomycete Genus Salinispora: A Model Organism for Secondary Metabolite Discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef]
- Le Loarer, A.; Marcellin-Gros, R.; Dufossé, L.; Bignon, J.; Frédérich, M.; Ledoux, A.; Queiroz, E.F.; Wolfender, J.-L.; Gauvin-Bialecki, A.; Fouillaud, M. Prioritization of Microorganisms Isolated from the Indian Ocean Sponge Scopalina Hapalia Based on Metabolomic Diversity and Biological Activity for the Discovery of Natural Products. Microorganisms 2023, 11, 697. [Google Scholar] [CrossRef]
- Schmid, R.; Petras, D.; Nothias, L.-F.; Wang, M.; Aron, A.T.; Jagels, A.; Tsugawa, H.; Rainer, J.; Garcia-Aloy, M.; Dührkop, K.; et al. Ion Identity Molecular Networking for Mass Spectrometry-Based Metabolomics in the GNPS Environment. Nat. Commun. 2021, 12, 3832. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Rutz, A.; Sorokina, M.; Galgonek, J.; Mietchen, D.; Willighagen, E.; Gaudry, A.; Graham, J.G.; Stephan, R.; Page, R.; Vondrášek, J.; et al. The LOTUS Initiative for Open Knowledge Management in Natural Products Research. eLife 2022, 11, e70780. [Google Scholar] [CrossRef] [PubMed]
- Rutz, A.; Dounoue-Kubo, M.; Ollivier, S.; Bisson, J.; Bagheri, M.; Saesong, T.; Ebrahimi, S.N.; Ingkaninan, K.; Wolfender, J.-L.; Allard, P.-M. Taxonomically Informed Scoring Enhances Confidence in Natural Products Annotation. Front. Plant Sci. 2019, 10, 1329. [Google Scholar] [CrossRef] [PubMed]
- Le Goff, G.; Adelin, E.; Cortial, S.; Servy, C.; Ouazzani, J. Application of Solid-Phase Extraction to Agar-Supported Fermentation. Bioprocess. Biosyst. Eng. 2013, 36, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Farinella, V.F.; Kawafune, E.S.; Tangerina, M.M.P.; Domingos, H.V.; Costa-Lotufo, L.V.; Ferreira, M.J.P. OSMAC Strategy Integrated with Molecular Networking for Accessing Griseofulvin Derivatives from Endophytic Fungi of Moquiniastrum Polymorphum (Asteraceae). Molecules 2021, 26, 7316. [Google Scholar] [CrossRef] [PubMed]
- Marquez, J.; Murawski, A.; Wagman, G.H.; Jaret, R.S.; Reimann, H. Isolation, purification and preliminary characterization of megalomicin. J. Antibiot. 1969, 22, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Useglio, M.; Peirú, S.; Rodríguez, E.; Labadie, G.R.; Carney, J.R.; Gramajo, H. TDP-l-Megosamine Biosynthesis Pathway Elucidation and Megalomicin A Production in Escherichia coli. Appl. Environ. Microbiol. 2010, 76, 3869. [Google Scholar] [CrossRef]
- MetaCyc: Metabolic Pathways from All Domains of Life. Available online: https://metacyc.org/ (accessed on 27 July 2023).
- Goodman, C.D.; Useglio, M.; Peirú, S.; Labadie, G.R.; McFadden, G.I.; Rodríguez, E.; Gramajo, H. Chemobiosynthesis of New Antimalarial Macrolides. Antimicrob. Agents Chemother. 2013, 57, 907–913. [Google Scholar] [CrossRef]
- Armin, R.; Zühlke, S.; Grunewaldt-Stöcker, G.; Mahnkopp-Dirks, F.; Kusari, S. Production of Siderophores by an Apple Root-Associated Streptomyces Ciscaucasicus Strain GS2 Using Chemical and Biological OSMAC Approaches. Molecules 2021, 26, 3517. [Google Scholar] [CrossRef]
- Tanaka, M.; Shin-Ya, K.; Furihata, K.; Seto, H. Isolation and Structural Elucidation of Antioxidative Substances, Carbazoquinocins A to F. J. Antibiot. 1995, 48, 326–328. [Google Scholar] [CrossRef]
- Kashiwada, Y.; Nonaka, G.; Nishioka, I. Studies on Rhubarb (Rhei Rhizoma). V. Isolation and Characterization of Chromone and Chromanone Derivatives. Chem. Pharm. Bull. 1984, 32, 3493–3500. [Google Scholar] [CrossRef]
- Yeo, W.-H.; Yun, B.-S.; Kim, S.-S.; Park, E.-K.; Kim, Y.-H.; Yoo, I.-D.; Yu, S.-H. GTRI-02, a New Lipid Peroxidation Inhibitor from Micromonospora Sp. SA246. J. Antibiot. 1998, 51, 952–953. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.M.; Masand, N.; Verma, S.; Masand, V. Chromones: Privileged Scaffold in Anticancer Drug Discovery. Chem. Bio. Drug Des. 2021, 98, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Liu, D.; Huo, X.; Qiu, D. The Anticancer Activity of Carbazole Alkaloids. Arch. Pharm. 2022, 355, 2100277. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, N.; Tanaka, Y.; Kawamoto, T. Aristeromycin and DZNeP Cause Growth Inhibition of Prostate Cancer via Induction of Mir-26a. Eur. J. Pharmacol. 2017, 812, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Crüsemann, M.; O’Neill, E.C.; Larson, C.B.; Melnik, A.V.; Floros, D.J.; da Silva, R.R.; Jensen, P.R.; Dorrestein, P.C.; Moore, B.S. Prioritizing Natural Product Diversity in a Collection of 146 Bacterial Strains Based on Growth and Extraction Protocols. J. Nat. Prod. 2017, 80, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Williams, P.G.; Oh, D.-C.; Zeigler, L.; Fenical, W. Species-Specific Secondary Metabolite Production in Marine Actinomycetes of the Genus Salinispora. Appl. Environ. Microbiol. 2007, 73, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Velasco-Alzate, K.; Dias, T.; Macedo, H.; Ferreira, E.G.; Jimenez, P.C.; Lotufo, T.M.C.; Lopes, N.P.; Gaudêncio, S.P.; Costa-Lotufo, L.V. Metabolomic Fingerprinting of Salinispora From Atlantic Oceanic Islands. Front. Microbiol. 2018, 9, 3021. [Google Scholar] [CrossRef] [PubMed]
- Schlawis, C.; Harig, T.; Ehlers, S.; Guillen-Matus, D.G.; Creamer, K.E.; Jensen, P.R.; Schulz, S. Extending the Salinilactone Family. Chembiochem 2020, 21, 1629–1632. [Google Scholar] [CrossRef]
- Stratmann, A.; Schupp, T.; Toupet, C.; Schilling, W.; Oberer, L.; Traber, R. New Insights into Rifamycin B Biosynthesis: Isolation of Proansamycin B and 34a-Deoxy-Rifamycin W as Early Macrocyclic Intermediates Indicating Two Separated Biosynthetic Pathways. J. Antibiot. 2002, 55, 396–406. [Google Scholar] [CrossRef]
- Jimenez, P.C.; Wilke, D.V.; Ferreira, E.G.; Takeara, R.; de Moraes, M.O.; da Silveira, E.R.; da Cruz Lotufo, T.M.; Lopes, N.P.; Costa-Lotufo, L.V. Structure Elucidation and Anticancer Activity of 7-Oxostaurosporine Derivatives from the Brazilian Endemic Tunicate Eudistoma Vannamei. Mar. Drugs 2012, 10, 1092–1102. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Li, H.; Xu, M.; Li, T.; Wang, J.; Sun, C.; Hong, K.; Li, W. Staurosporine Derivatives Generated by Pathway Engineering in a Heterologous Host and Their Cytotoxic Selectivity. J. Nat. Prod. 2018, 81, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching Molecular Structure Databases with Tandem Mass Spectra Using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic Classification of Unknown Metabolites Using High-Resolution Fragmentation Mass Spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Chem. 2016, 8, 61. [Google Scholar] [CrossRef]
- Trager, W.; Jensen, J.B. Human Malaria Parasites in Continuous Culture. Science 1976, 193, 673–675. [Google Scholar] [CrossRef]









| Compound ID | Molecular Formula | Compound Name or InChIKey (1,2,3) | Chemical Class | Similarity (1,2,3) |
|---|---|---|---|---|
| C82.1 | C44H80N2O15 | Megalomicin A (1,3) | Organooxygen compounds | 92% (1)/0.05 (3) |
| C82.2 | C45H78N2O17 | Megalomicin B (1,3) | 89% (1)/0.05 (3) | |
| C82.3 | C47H84N2O16 | 4′-Propionylmegalomicin A (1) | 89% (1) | |
| C82.4 | C48H84N2O17 | Megalomicin C1 (1,3) | 87% (1)/0.05 (3) | |
| C82.5 | C49H86N2O17 | Megalomicin C2 (1,3) | 84% (1)/0.05 (3) | |
| C82.6 | C39H69NO14 | 2′-O-Acetylerythromycin A (1) | 83% (1) | |
| C82.7 | C36H65NO13 | Erythromycin C (1) | 98% (1) | |
| C82.8 | 13-Deethyl-13-methylerythromycin (1) | 96% (1) | ||
| C82.9 | C36H65NO12 | Erythromycin D (1) | 93% (1) | |
| C82.10 | C37H67NO12 | Erythromycin B (1,3) | 91% (1)/0.05 (3) | |
| C82.11 | C36H63NO12 | 6-Desmethyl erythromycin D (1,3) | 92% (1)/0.05 (3) | |
| C82.12 | C29H53NO9 | 3-O-De(3-C,3-O-dimethyl-2,6-dideoxy-alpha-L-ribo-hexopyranosyl)-6-deoxyerythromycin (1) | 85.3% (1) | |
| C82.13 | ||||
| C82.14 | C36H65NO12 | 6-Deoxy-3′-O-demethylerythromycin (1) | 89% (1) | |
| C82.15 | C35H63NO13 | Norerythromycin (1) | 89% (1) | |
| C82.16 | C21H38O7 | Erythronolide B (1) | Macrolides and analogues | 62% (1) |
| C82.17 | C20H36O7 | 2-Desmethyl-2-hydroxy-6-deoxyerythronolide B (1) | 69% (1) | |
| C82.18 | C28H50O10 | 3-O-Alpha-mycarosylerythronolide B (1) | 68% (1) | |
| C82.19 | C27H48O10 | 3-O-(alpha-L-olivosyl)erythronolide B (1) | 75% (1) | |
| C82.20 | 75% (1) | |||
| C82.21 | C21H38O6 | 6-Deoxyerythronolide B (1,3) | 63% (1)/0.05 (3) |
| Compound ID | Molecular Formula | Compound Name or InChIKey (1,2,3) | Chemical Class | Similarity (1,2,3) |
|---|---|---|---|---|
| C57.1 | C11H15N5O3 | 2-Deoxy-N6-methyladenosine (1) | Purine nucleoside | 93% (1) |
| C57.2 | C11H15N5O | SXIDRQQQIPLCTJ (1) | 81% (1) | |
| C57.3 | C11H13N5O3 | UHYRJPGYRFMFLT (1) | 75% (1) | |
| C57.4 | C11H15N5 | 9-cyclopentyl-N-methylpurin-6-amine (1) | 75% (1) | |
| C57.5 | C12H17N5OS | INPAYTORGXXLMB (1) | 66% (1) | |
| C57.6 | C14H26N2O5S | JDNYVZBVEBRRCT (1) | Carboxylic acids and derivatives | 86% (1) |
| C57.7 | C13H24NO5S | (?)-S-Acetylpantetheine (1) | 77% (1) | |
| C57.8 | C17H30N2O5S | ZCNIMMSEOJFZKZ (1) | 81% (1) | |
| C57.9 | C19H28N2O5S | IXKOTSUCYPEFPP (1) | 78% (1) | |
| C57.10 | C9H9NO | Indole-3-carbinol (1,2) | Indoles and derivatives | 88% (1)/0.77 (2) |
| C57.11 | ||||
| C57.12 | C13H12O4 | Aloesone (1) | Benzopyrans | 71% (1) |
| C57.13 | C13H14O4 | Aloesol (1) | 83% (1) | |
| C57.14 | C20H23NO2 | Carbazoquinocin C (1,3) | Indoles and derivatives | 71% (1)/0.05 (3) |
| C57.15 | C21H25NO2 | Carbazoquinocin E (1,3) | 61% (1)/0.05 (3) | |
| C57.16 | C22H27NO2 | Carbazoquinocin F (1,3) | 58% (1)/0.05 (3) | |
| C57.17 | C24H31NO2 | 12-Carbazol-9-yldodecanoic acid (1) | 53% (1) |
| Compound ID | Molecular Formula | Compound Name or InChIKey (1,2,3) | Chemical Class | Similarity (1,2,3) |
|---|---|---|---|---|
| C78.1 | C28H26N4O4 | OH staurosporine (1,2,3) | Indoles and derivatives | 85% (1)/0.79 (2)/0.1 (3) |
| C78.2 | C28H26N4O3 | Staurosporine (1,2,3) | 98% (1)/0.96 (2)/0.1 (3) | |
| C78.3 | C29H28N4O4 | 4′-N-methyl-5′-hydroxy-staurosporine (1) | 78% (1) | |
| C78.4 | C28H24N4O5 | 4′-demethyl-Af-formyl-7V-hydroxy-staurosporine (1) | 74% (1) | |
| C78.5 | C37H45NO12 | Rifamycin S (1,2,3) | Macrolactams | 75% (1)/0.74 (2)/0.05 (3) |
| C78.6 | C36H43NO12 | 16-demethyl rifamycin S (3) | 0.1 (3) | |
| C78.7 | C35H45NO10 | 34a-deoxy-rifamycin W (1) | 62% (1) | |
| C78.8 | 58% (1) | |||
| C78.9 | C34H41NO10 | Proansamycin B (1,3) | 64% (1)/0.05 (3) | |
| C78.10 | 54%(1) | |||
| C78.11 | C34H41NO11 | Demethyl-desacetyl-rifamycin S (1) | 71% (1) | |
| C78.12 | C37H45NO13 | 20-hydroxyrifamycin S (1,3) | 74% (1)/0.05 (3) | |
| C78.13 | C35H42NO12 | 30-hydroxyrifamycin W (1) | 59% (1) | |
| C78.14 | C22H37NO5 | Saliniketal A (1,3) | Prenol lipids | 43% (1)/0.05 (3) |
| C78.15 | C22H37NO6 | Saliniketal B (1,3) | 49% (1)/0.13 (3) | |
| C78.16 | C8H10O3 | Salinilactone D (3) | Lactones | 0.13 (3) |
| C78.17 | C9H12O3 | Salinilactone E (3) | 0.13 (3) | |
| C78.18 | 0.13 (3) | |||
| C78.19 | C10H14O3 | Salinilactone A (3) | 0.13 (3) | |
| C78.20 | 0.13 (3) | |||
| C78.21 | 0.13 (3) | |||
| C78.22 | C11H16O3 | Salinilactone C (3) | 0.13 (3) | |
| C78.23 | C12H18O3 | Salinilactone H (3) | 0.13 (3) |
| Microbial Strains | Days | Support | Culture Medium |
|---|---|---|---|
| Micromonospora sp. (SH-82 and SH-57) | 7/14/21 | Solid/Liquid | A1BFe+C |
| 14 | MB | ||
| Salinispora arenicola SH-78 | 7/14/21 | Solid | A1BFe+C |
| 14 | Liquid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Loarer, A.; Dufossé, L.; Bignon, J.; Frédérich, M.; Ledoux, A.; Fouillaud, M.; Gauvin-Bialecki, A. OSMAC Method to Assess Impact of Culture Parameters on Metabolomic Diversity and Biological Activity of Marine-Derived Actinobacteria. Mar. Drugs 2024, 22, 23. https://doi.org/10.3390/md22010023
Le Loarer A, Dufossé L, Bignon J, Frédérich M, Ledoux A, Fouillaud M, Gauvin-Bialecki A. OSMAC Method to Assess Impact of Culture Parameters on Metabolomic Diversity and Biological Activity of Marine-Derived Actinobacteria. Marine Drugs. 2024; 22(1):23. https://doi.org/10.3390/md22010023
Chicago/Turabian StyleLe Loarer, Alexandre, Laurent Dufossé, Jérôme Bignon, Michel Frédérich, Allison Ledoux, Mireille Fouillaud, and Anne Gauvin-Bialecki. 2024. "OSMAC Method to Assess Impact of Culture Parameters on Metabolomic Diversity and Biological Activity of Marine-Derived Actinobacteria" Marine Drugs 22, no. 1: 23. https://doi.org/10.3390/md22010023
APA StyleLe Loarer, A., Dufossé, L., Bignon, J., Frédérich, M., Ledoux, A., Fouillaud, M., & Gauvin-Bialecki, A. (2024). OSMAC Method to Assess Impact of Culture Parameters on Metabolomic Diversity and Biological Activity of Marine-Derived Actinobacteria. Marine Drugs, 22(1), 23. https://doi.org/10.3390/md22010023