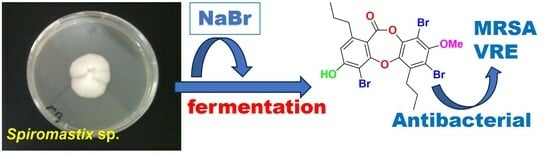
Brominated Depsidones with Antibacterial Effects from a Deep-Sea-Derived Fungus Spiromastix sp.
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation and Extraction
3.4. Isolation and Purification
3.5. Antibacterial Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agarwal, V.; Miles, Z.D.; Winter, J.M.; Eustaquio, A.S.; El Gamal, A.A.; Moore, B.S. Enzymatic halogenation and dehalogenation reactions: Pervasive and mechanistically diverse. Chem. Rev. 2017, 117, 5619–5674. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.M.; Kannan, R.; Kopecka, H.; Harris, T.M. The role of the chlorine substituents in the antibiotic vancomycin: Preparation and characterization of mono- and didechlorovancomycin. J. Am. Chem. Soc. 1985, 107, 6652–6658. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, Z.; Liu, Y.; Lu, Y.; Chen, K.; Zhu, W. Halogen bond: Its role beyond drug-target binding affinity for drug discovery and development. J. Chem. Inf. Model. 2014, 54, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, P. The unique role of halogen substituents in the design of modern agrochemicals. Pest Manag. Sci. 2010, 66, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.R.; Peterson, B.L.; Appelbaum, F.R.; Kolitz, J.; Elias, L.; Shepherd, L.; Hines, J.; Threatte, G.A.; Larson, R.A.; Cheson, B.D.; et al. Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia. N. Engl. J. Med. 2000, 343, 1750–1757. [Google Scholar] [CrossRef] [PubMed]
- Prudhomme, M. Recent developments of rebeccamycin analogues as topoisomerase I inhibitors and antitumor agents. Curr. Med. Chem. 2000, 7, 1189–1212. [Google Scholar] [CrossRef] [PubMed]
- Fraley, A.E.; Sherman, D.H. Halogenase engineering and its utility in medicinal chemistry. Bioorg. Med. Chem. Lett. 2018, 28, 1992–1999. [Google Scholar] [CrossRef]
- Wang, J.; Pang, X.; Chen, C.; Gao, C.; Zhou, X.; Liu, Y.; Luo, X. Chemistry, biosynthesis, and biological activity of halogenated compounds produced by marine microorganisms. Chin. J. Chem. 2022, 40, 1729–1750. [Google Scholar] [CrossRef]
- Molchanova, N.; Nielsen, J.E.; Sørensen, K.B.; Prabhala, B.K.; Hansen, P.R.; Lund, R.; Barron, A.E.; Jenssen, H. Halogenation as a tool to tune antimicrobial activity of peptoids. Sci. Rep. 2020, 10, 14805. [Google Scholar] [CrossRef]
- Bister, B.; Bischoff, D.; Nicholson, G.J.; Stockert, S.; Wink, J.; Brunati, C.; Donadio, S.; Pelzer, S.; Wohlleben, W.; Süssmuth, R.D. Bromobalhimycin and chlorobromobalhimycins—Illuminating the potential of halogenases in glycopeptide antibiotic biosyntheses. ChemBioChem 2003, 4, 658–662. [Google Scholar] [CrossRef]
- Niu, S.; Liu, D.; Hu, X.; Proksch, P.; Shao, Z.; Lin, W. Spiromastixones A-O, antibacterial chlorodepsidones from a deep-sea-derived Spiromastix sp. fungus. J. Nat. Prod. 2014, 77, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Liu, D.; Shao, Z.; Huang, J.; Fan, A.; Lin, W. Chlorinated metabolites with antibacterial activities from a deep-sea-derived Spiromastix fungus. RSC Adv. 2021, 11, 29661–29667. [Google Scholar] [CrossRef]
- Niu, S.; Liu, D.; Proksch, P.; Shao, Z.; Lin, W. New polyphenols from a deep sea Spiromastix sp. fungus, and their antibacterial activities. Mar. Drugs 2015, 13, 2526–2540. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Si, L.; Liu, D.; Zhou, A.; Zhang, Z.; Shao, Z.; Wang, S.; Zhang, L.; Zhou, D.; Lin, W. Spiromastilactones: A new class of influenza virus inhibitors from deep-sea fungus. Eur. J. Med. Chem. 2016, 108, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.K.; Zhu, W.Y.; Zhao, L.X.; Chen, Y.C.; Li, S.J.; Cheng, P.; Ge, H.M.; Tan, R.X.; Jiao, R.H. New antibacterial depsidones from an ant-derived fungus Spiromastix sp. MY-1. Chin. J. Nat. Med. 2022, 20, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhou, Z.; Chen, Y.; Song, Y.; Ju, J. Characterization of the depsidone gene cluster reveals etherification, decarboxylation and multiple halogenations as tailoring steps in depsidone assembly. Acta Pharm. Sin. B 2023, 13, 3919–3929. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, D.; Gao, S.; Cai, X.; Yao, M.; Xu, Y.; Gong, Y.; Zheng, K.; Mao, Y.; Yang, L.; et al. Didepside formation by the nonreducing polyketide synthase Preu6 of preussia isomera requires interaction of starter acyl transferase and thioesterase domains. Angew. Chem. Int. Ed. 2023, 62, e202214379. [Google Scholar] [CrossRef]
- Chen, L.; Wei, X.; Matsuda, Y. Depside bond formation by the starter-unit acyltransferase domain of a fungal polyketide synthase. J. Am. Chem. Soc. 2022, 144, 19225–19230. [Google Scholar] [CrossRef]
- Larsen, J.; Raisen, C.L.; Ba, X.; Sadgrove, N.J.; Padilla-Gonzalez, G.F.; Simmonds, M.S.J.; Loncaric, I.; Kerschner, H.; Apfalter, P.; Hartl, R.; et al. Emergence of methicillin resistance predates the clinical use of antibiotics. Nature 2022, 602, 135–141. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- Nelson, R.E.; Hatfield, K.M.; Wolford, H.; Samore, M.H.; Scott, R.D.; Reddy, S.C.; Olubajo, B.; Paul, P.; Jernigan, J.A.; Baggs, J. National estimates of healthcare costs associated with multidrug-resistant bacterial infections among hospitalized patients in the United States. Clin. Infect. Dis. 2021, 72, S17–S26. [Google Scholar] [CrossRef]
- Chen, S.; Liu, D.; Zhang, Q.; Guo, P.; Ding, S.; Shen, J.; Zhu, K.; Lin, W. A marine antibiotic kills multidrug-resistant bacteria without detectable high-level resistance. ACS Infect. Dis. 2021, 7, 884–893. [Google Scholar] [CrossRef] [PubMed]


| Position | 1 a | 2 a | 3 a | 4 a | 5 a | 6 b | 7 a | 8 a | 9 b | 10 a | 11 c |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 6.80, s | 6.71, s | 6.84, s | ||||||||
| 5 | 6.79, s | 6.80, s | 6.84, s | ||||||||
| 8 | 2.81, t (7.9) | 2.66, t (7.8) | 2.70, t (7.6) | 2.86, t (7.7) | 3.11, t (8.0) | 2.78, t (7.9) | 2.68, t (7.7) | 2.85, m | 2.81, t (7.7) | 2.79, m | 2.72, t (8.0) |
| 9 | 1.54, m | 1.49, m | 1.49, m | 1.54, m | 1.59, m | 1.47, m | 1.48, m | 1.55, m | 1.58, m | 1.53, m | 1.56, m |
| 10 | 0.89, t (7.3) | 0.86, t (7.3) | 0.83, t (7.2) | 0.86, t (7.3) | 0.88, t (7.3) | 0.90, t (7.3) | 0.83, t (7.3) | 0.88, t (7.2) | 0.87, t (7.3) | 0.90, t (7.2) | 0.87, t (7.2) |
| 3′ | 6.50, d (2.7) | 6.77, s | 6.80, s | 7.12, s | |||||||
| 5′ | 6.47, d (2.7) | 6.69, s | |||||||||
| 6′ | 7.07, s | ||||||||||
| 7′ | 2.65, t (7.8) | 3.13, t (8.0) | 2.90, t (7.8) | 2.81, t (8.1) | 2.76, t (7.8) | 3.14, t (7.5) | 3.16, t (8.0) | 2.87, m | 3.15, t (7.9) | 2.81, m | 2.83, t (7.5) |
| 8′ | 1.57, m | 1.48, m | 1.07, m | 1.51, m | 1.48, m | 1.61, m | 1.50, m | 1.55, m | 1.50, m | 1.50, m | 1.54, m |
| 9′ | 0.97, t (7.3) | 1.00, t (7.3) | 0.95, t (7.2) | 0.98, t (7.3) | 1.01, t (7.3) | 1.01, t (7.3) | 1.00, t (7.2) | 1.04, t (7.2) | 1.02, t (7.3) | 0.90, t (7.2) | 0.92, t (7.2) |
| OMe | 3.84, s | 3.78, s | 3.76, s | 3.78, s | 3.85, s | 3.78, s |
| Position | 1 a | 2 a | 3 a | 4 a | 5 a | 6 b | 7 a | 8 a | 9 b | 10 a | 11 c |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 111.1, C | 112.5, C | 112.3, C | 112.9, C | 113.6, C | 113.2, C | 111.5, C | 111.7, C | 112.7, C | 112.6, C | - |
| 2 | 161.7, C | 160.4, C | 160.6, C | 161.7, C | 159.3, C | 159.2, C | 160.2, C | 161.0, C | 158.8, C | 157.6, C | 158.9, C |
| 3 | 105.3, CH | 98.9, C | 99.0, C | 105.2, CH | 101.3, C | 101.3, C | 99.2, C | 105.7, CH | 101.5, C | 102.5, C | 102.2, C |
| 4 | 159.7, C | 159.6, C | 159.7, C | 161.7, C | 159.3, C | 159.2, C | 160.0, C | 161.0, C | 158.8, C | 157.6, C | 158.9, C |
| 5 | 113.6, C | 115.1, CH | 115.3, C | 110.3, C | 112.8, C | 114.1, C | 115.6, C | 112.4, C | 113.8, C | 114.1, C | 116.2, C |
| 6 | 146.9, C | 148.5, C | 148.1, C | 146.9, C | 145.8, C | 146.1, C | 148.8, C | 147.3, C | 146.1, C | 144.6, C | 145.7, C |
| 7 | 162.9, C | 162.1, C | 161.8, C | 161.9, C | 162.0, C | 161.7, C | 160.7, C | 161.2, C | 160.6, C | 161.9, C | 161.6, C |
| 8 | 36.0, CH2 | 35.7, CH2 | 35.5, CH2 | 35.7, CH2 | 36.9, CH2 | 27.1, CH2 | 35.7, CH2 | 35.7, CH2 | 36.6, CH2 | 36.1, CH2 | 36.9, CH2 |
| 9 | 23.0, CH2 | 24.5, CH2 | 24.6, CH2 | 22.9, CH2 | 22.9, CH2 | 23.1, CH2 | 24.5, CH2 | 22.8, CH2 | 22.7, CH2 | 23.0, CH2 | 23.0, CH2 |
| 10 | 14.3, CH3 | 14.3, CH3 | 14.1, CH3 | 14.2, CH3 | 14.3, CH3 | 14.4, CH3 | 14.2, CH3 | 14.2, CH3 | 14.1, CH3 | 14.1, CH3 | 14.5, CH3 |
| 1′ | 141.3, C | 142.0, C | 142.7, C | 142.0, C | 142.0, C | 143.0, C | 146.7, C | 146.1, C | 146.6, C | 150.1, C | 149.1, C |
| 2′ | 144.6, C | 143.4, C | 142.9, C | 142.1, C | 143.4, C | 143.7, C | 142.2, C | 142.5, C | 142.2, C | 136.5, C | 141.9, C |
| 3′ | 105.5, CH | 105.6, CH | 99.5, C | 102.2, C | 105.6, C | 103.5, CH | 108.3, C | 108.0, C | 108.3, C | 135.2, C | 134.0, C |
| 4′ | 155.2, C | 152.9, C | 153.0, C | 151.0, C | 153.0, C | 154.4, C | 152.8, C | 152.5, C | 152.9, C | 110.8, C | 117.5, C |
| 5′ | 113.2, CH | 108.8, C | 113.3, CH | 111.1, C | 108.9, C | 110.0, C | 117.2, C | 116.6, C | 117.2, C | 153.9, C | 152.0, C |
| 6′ | 135.8, C | 136.0, C | 134.8, C | 134.0, C | 135.9, C | 136.1, C | 135.5, C | 135.2, C | 135.5, C | 103.6, C | 108.3, C |
| 7′ | 31.4, CH2 | 33.0, CH2 | 32.7, CH2 | 32.6, CH2 | 33.0, CH2 | 33.0, CH2 | 33.2, CH2 | 32.5, CH2 | 33.2, CH2 | 32.3, CH2 | 32.5, CH2 |
| 8′ | 23.7, CH2 | 23.1, CH2 | 24.2, CH2 | 22.5, CH2 | 23.1, CH2 | 22.9, CH2 | 22.8, CH2 | 22.5, CH2 | 22.9, CH2 | 22.1, CH2 | 22.0, CH2 |
| 9′ | 14.4, CH3 | 14.4, CH3 | 14.1, CH3 | 14.9, CH3 | 14.4, CH3 | 14.4, CH3 | 14.3, CH3 | 14.5, CH3 | 14.3, CH3 | 14.4, CH3 | 14.0, CH3 |
| OMe | 57.6, CH3 | 60.9, CH3 | 60.9, CH3 | 60.8, CH3 | 57.3, CH3 | 60.8, CH3 |
| Compounds | MRSA T144 | MRSA 1530 | VRE CAU360 | VRE CAU378 | E. coli ATCC 25922 | E. coli B2 (mcr-1 + blaNDM-5) |
|---|---|---|---|---|---|---|
| 1 | 32 | 32 | 64 | 64 | >64 | >64 |
| 2 | 8 | 8 | 8 | 8 | >64 | >64 |
| 3 | 16 | 16 | 32 | 32 | >64 | >64 |
| 4 | 4 | 8 | 16 | 16 | >64 | >64 |
| 5 | 4 | 4 | 8 | 8 | >64 | >64 |
| 6 | 2 | 2 | 2 | 2 | >64 | >64 |
| 7 | 0.5 | 0.5 | 1 | 1 | >64 | >64 |
| 8 | 1 | 1 | 2 | 2 | >64 | >64 |
| 9 | 2 | 2 | 1 | 1 | >64 | >64 |
| 10 | 2 | 2 | 2 | 2 | >64 | >64 |
| 11 | 8 | 8 | 4 | 4 | >64 | >64 |
| 12 | 8 | 8 | 8 | 8 | >64 | >64 |
| 13 | 16 | 16 | 16 | 16 | >64 | >64 |
| 14 | 64 | 64 | 64 | 64 | >64 | >64 |
| 15 | 64 | 64 | >64 | >64 | >64 | >64 |
| 16 | 2 | 2 | 2 | 2 | >64 | >64 |
| Spiromastixone J | 2 | 2 | 2 | 2 | >64 | >64 |
| Vancomycin * | 0.35 | 0.35 | / | / | / | / |
| Linezolid * | / | / | 5.93 | 11.86 | / | / |
| Colistin * | / | / | / | / | 0.22 | 6.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Liu, D.; Chen, S.; Ren, J.; Gao, C.; Li, Z.; Fan, A.; Lin, W. Brominated Depsidones with Antibacterial Effects from a Deep-Sea-Derived Fungus Spiromastix sp. Mar. Drugs 2024, 22, 78. https://doi.org/10.3390/md22020078
Huang Z, Liu D, Chen S, Ren J, Gao C, Li Z, Fan A, Lin W. Brominated Depsidones with Antibacterial Effects from a Deep-Sea-Derived Fungus Spiromastix sp. Marine Drugs. 2024; 22(2):78. https://doi.org/10.3390/md22020078
Chicago/Turabian StyleHuang, Zequan, Dong Liu, Shang Chen, Jinwei Ren, Chenghai Gao, Zhiyong Li, Aili Fan, and Wenhan Lin. 2024. "Brominated Depsidones with Antibacterial Effects from a Deep-Sea-Derived Fungus Spiromastix sp." Marine Drugs 22, no. 2: 78. https://doi.org/10.3390/md22020078
APA StyleHuang, Z., Liu, D., Chen, S., Ren, J., Gao, C., Li, Z., Fan, A., & Lin, W. (2024). Brominated Depsidones with Antibacterial Effects from a Deep-Sea-Derived Fungus Spiromastix sp. Marine Drugs, 22(2), 78. https://doi.org/10.3390/md22020078