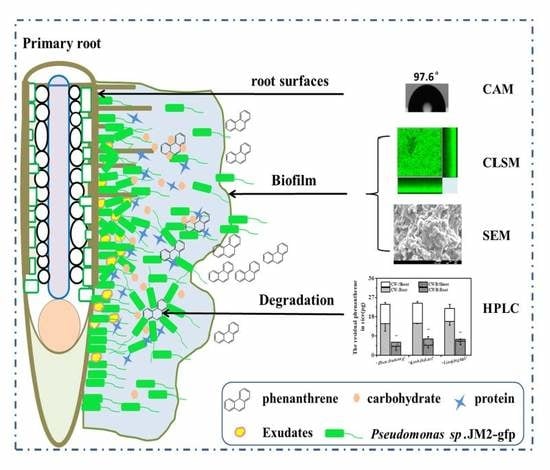
Characterization of Biofilm Formed by Phenanthrene-Degrading Bacteria on Rice Root Surfaces for Reduction of PAH Contamination in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plants Preparation
2.3. Root Surfaces Characterization
2.3.1. Scanning Electron Microscope (SEM)
2.3.2. Water Contact Angle
2.3.3. Initial Adhesion Measurement
2.4. Biofilm Formation
2.5. Characterization of the Biofilm on Root Surfaces
2.5.1. Scanning Electron Microscope (SEM)
2.5.2. Confocal Laser Scanning Microscopy (CLSM)
2.5.3. Composite Extracellular Polymeric Substances (EPS) Characterization
2.6. Degradation of Phenanthrene by the Biofilm
2.7. Statistics
3. Results and Discussion
3.1. Morphology of Rice Root Surfaces
3.2. Hydrophobicity Characterization
3.3. Initial Adhesion of Rice Root Surface
3.4. Biofilm Formation on the Root Surfaces
3.5. Characterization of Biofilm on the Root Surfaces
3.5.1. Morphology of biofilms
3.5.2. Characterization of Bacterial Biofilm
3.5.3. Characterization of Biofilm Associated EPS
3.6. Capacity of the Phenanthrene Degradation by the Biofilm on Rice Root Surfaces
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Sun, J.; Leu, S.Y.; Chen, S. Toward a fundamental understanding of cellulase-lignin interactions in the whole slurry enzymatic saccharification process: Elucidating Cellulase-Lignin Interactions for Whole Slurry Saccharification. Biofuels Bioprod. Biorefin. 2016, 10, 648–663. [Google Scholar] [CrossRef]
- Xu, N.N.; Bao, M.T.; Sun, P.Y.; Li, Y.M. Study on bioadsorption and biodegradation of petroleum hydrocarbons by a microbial consortium. Bioresour. Technol. 2013, 149, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Xu, L.; Jia, L.Y. Characterization of pyrene degradation by Pseudomonas sp. strain Jpyr-1 isolated from active sewage sludge. Bioresour. Technol. 2013, 140, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kotoky, R.; Das, S.; Singha, L.P.; Pandey, P.; Singha, K.M. Biodegradation of Benzo(a)pyrene by biofilm forming and plant growth promoting Acinetobacter sp. strain PDB4. Environ. Technol. Innov. 2017, 8, 256–268. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Z.M.; Sheng, Y.H.; Gao, Y.Z.; Zhao, Z.H. Phenanthrene-degrading bacteria on root surfaces: A natural defense that protects plants from phenanthrene contamination. Plant. Soil 2018, 425, 335–350. [Google Scholar] [CrossRef]
- Beykal, B.; Herzberg, M.; Oren, Y.; Mauter, M.S. Influence of surface charge on the rate, extent, and structure of adsorbed Bovine Serum Albumin to gold electrodes. J. Colloid Interf. Sci. 2015, 460, 321–328. [Google Scholar] [CrossRef]
- Pandit, S.; Shanbhag, S.; Mauter, M.S.; Oren, Y.; Herzberg, M. The influence of electric fields on biofouling of carbonaceous electrodes. Environ. Sci. Technol. 2017, 51. [Google Scholar] [CrossRef]
- Tribedi, P.; Sil, A.K. Cell surface hydrophobicity: A key component in the degradation of polyethylene succinate by Pseudomonas sp. AKS2. J. Appl. Microbiol. 2014, 116, 295–303. [Google Scholar] [CrossRef]
- Garrett, T.R.; Bhakoo, M.; Zhang, Z.B. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Simes, M.; Simoes, L.C.; Cleto, S.; Pereira, M.O.; Vieira, M.J. The effects of a biocide and a surfactant on the detachment of Pseudomonas fluorescens from glass surfaces. Int. J. Food Microbiol. 2008, 121, 335–341. [Google Scholar] [CrossRef] [Green Version]
- Sinde, E.; Carballo, J. Attachment of Salmonella spp. and Listeria monocytogenes to stainless steel, rubber and polytetrafluorethylene: The influence of free energy and the effect of commercial sanitizers. Food Microbiol. 2000, 17, 439–447. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Cunliffe, D.; Smart, C.A.; Alexander, C.; Vulfson, E.N. Bacterial adhesion at synthetic surfaces. Appl. Environ. Microbiol. 1999, 65, 4995–5002. [Google Scholar]
- De Oliveira, D.C.V.; Fernandes, A.; Kaneno, R.; Silva, M.G.; Araujo, J.P.; Silva, N.C.C.; Rall, V.L.M. Ability of Salmonella spp. to Produce Biofilm Is Dependent on Temperature and Surface Material. Foodborne Pathog. Dis. 2014, 11, 478–483. [Google Scholar] [CrossRef]
- Rodriguez-Melcon, C.; Riesco-Pelaez, F.; Carballo, J.; Garcia-Fernandez, C.; Capita, R.; Alonso-Calleja, C. Structure and viability of 24-and 72-h-old biofilms formed by four pathogenic bacteria on polystyrene and glass contact surfaces. Food Microbiol. 2018, 76, 513–517. [Google Scholar] [CrossRef]
- Ben Abdallah, F.; Lagha, R.; Said, K.; Kallel, H.; Gharbi, J. Detection of Cell Surface Hydrophobicity, Biofilm and Fimbirae Genes in Salmonella Isolated from Tunisian Clinical and Poultry Meat. Iran. J. Public Health 2014, 43, 423–431. [Google Scholar]
- Dorosky, R.J.; Yu, J.M.; Pierson, L.S.; Pierson, E.A. Pseudomonas chlororaphis Produces Two Distinct R-Tailocins That Contribute to Bacterial Competition in Biofilms and on Roots. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Wu, K.; Yuan, S.F.; Xun, G.H.; Shi, W.; Pan, B.; Guan, H.L.; Shen, B.; Shen, Q.R. Root exudates from two tobacco cultivars affect colonization of Ralstonia solanacearum and the disease index. Eur. J. Plant. Pathol. 2015, 141, 667–677. [Google Scholar] [CrossRef]
- Beauregard, P.B.; Chai, Y.R.; Vlamakis, H.; Losick, R.; Kolter, R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. USA 2013, 110, E1621–E1630. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, D.Q.; Wang, D.D.; Miao, Y.Z.; Shao, J.H.; Zhou, X.; Xu, Z.H.; Li, Q.; Feng, H.C.; Li, S.Q.; et al. Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genom. 2015, 16, 685. [Google Scholar] [CrossRef]
- Schmidt, H.; Nunan, N.; Höck, A.; Eickhorst, T.; Kaiser, C.; Woebken, D.; Raynaud, X. Recognizing Patterns: Spatial Analysis of Observed Microbial Colonization on Root Surfaces. Front. Environ. Sci. 2018, 6, 61. [Google Scholar] [CrossRef]
- Eissenberger, K.; Moench, D.; Drissner, D.; Weiss, A.; Schmidt, H. Adherence factors of enterohemorrhagic Escherichia coli O157:H7 strain Sakai influence its uptake into the roots of Valerianella locusta grown in soil. Food Microbiol. 2018, 76, 245–256. [Google Scholar] [CrossRef]
- Ma, J.; Xu, L.; Jia, L.Y. Degradation of polycyclic aromatic hydrocarbons by Pseudomonas sp. JM2 isolated from active sewage sludge of chemical plant. J. Environ. Sci. 2012, 24, 2141–2148. [Google Scholar] [CrossRef]
- Jakoby, M.; Wang, H.Y.; Reidt, W.; Weisshaar, B.; Bauer, P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS. Lett. 2004, 577, 528–534. [Google Scholar] [CrossRef]
- Kaltenbach, R.; Diehl, D.; Schaumann, G.E. Links between nanoscale and macroscale surface properties of natural root mucilage studied by atomic force microscopy and contact angle. J. Colloid Interf. Sci. 2018, 516, 446–455. [Google Scholar] [CrossRef]
- Bonebrake, M.; Anderson, K.; Valiente, J.; Jacobson, A.; McLean, J.E.; Anderson, A.; Britt, D.W. Biofilms Benefiting Plants Exposed to ZnO and CuO Nanoparticles Studied with a Root-Mimetic Hollow Fiber Membrane. J. Agric. Food Chem. 2018, 66, 6619–6627. [Google Scholar] [CrossRef]
- Choudhry, P. High-Throughput Method for Automated Colony and Cell Counting by Digital Image Analysis Based on Edge Detection. PLoS ONE 2016, 11, e0148469. [Google Scholar] [CrossRef]
- Moreira, J.M.R.; Ponmozhi, J.; Campos, J.B.L.M.; Miranda, J.M.; Mergulhao, F.J. Micro- and macro-flow systems to study Escherichia coli adhesion to biomedical materials. Chem. Eng. Sci. 2015, 126, 440–445. [Google Scholar] [CrossRef]
- Nocelli, N.; Bogino, P.C.; Banchio, E.; Giordano, W. Roles of Extracellular Polysaccharides and Biofilm Formation in Heavy Metal Resistance of Rhizobia. Materials 2016, 9, 418. [Google Scholar] [CrossRef]
- Fujishige, N.A.; Kapadia, N.N.; De Hoff, P.L.; Hirsch, A.M. Investigations of Rhizobium biofilm formation. FEMS Microbiol. Ecol. 2006, 56, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersboll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef]
- Mangwani, N.; Shukla, S.K.; Kumari, S.; Rao, T.S.; Das, S. Characterization of Stenotrophomonas acidaminiphila NCW-702 biofilm for implication in the degradation of polycyclic aromatic hydrocarbons. J. Appl. Microbiol. 2014, 117, 1012–1024. [Google Scholar] [CrossRef]
- Mangwani, N.; Shukla, S.K.; Kumari, S.; Das, S.; Rao, T.S. Effect of biofilm parameters and extracellular polymeric substance composition on polycyclic aromatic hydrocarbon degradation. RSC Adv. 2016, 6, S7540–S7551. [Google Scholar] [CrossRef]
- Sun, K.; Liu, J.; Gao, Y.Z.; Jin, L.; Gu, Y.J.; Wang, W.Q. Isolation, plant colonization potential, and phenanthrene degradation performance of the endophytic bacterium Pseudomonas sp. Ph6-gfp. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef]
- Srey, S.; Jahid, I.K.; Ha, S.D. Biofilm formation in food industries: A food safety concern. Food Control. 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Renner, L.D.; Weibel, D.B. Physicochemical regulation of biofilm formation. MRS Bull. 2011, 36, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Bae, Y.M.; Lee, S.Y.; Lee, S.Y. Biofilm Formation of Staphylococcus aureus on Various Surfaces and Their Resistance to Chlorine Sanitizer. J. Food Sci. 2015, 80, M2279–M2286. [Google Scholar] [CrossRef]
- Sharma, S.; Jaimes-Lizcano, Y.A.; McLay, R.B.; Cirino, P.C.; Conrad, J.C. Subnanometric Roughness Affects the Deposition and Mobile Adhesion of Escherichia coli on Silanized Glass Surfaces. Langmuir 2016, 32, 5422–5433. [Google Scholar] [CrossRef]
- Olsson, A.L.J.; van der Mei, H.C.; Busscher, H.J.; Sharma, P.K. Novel Analysis of Bacterium-Substratum Bond Maturation Measured Using a Quartz Crystal Microbalance. Langmuir 2010, 26, 11113–11117. [Google Scholar] [CrossRef]
- Yu, P.; Wang, C.Y.; Zhou, J.L.; Jiang, L.; Xue, J.; Li, W. Influence of Surface Properties on Adhesion Forces and Attachment of Streptococcus mutans to Zirconia In Vitro. Biomed. Res. Int. 2016, 2016. [Google Scholar] [CrossRef]
- Harimawan, A.; Rajasekar, A.; Ting, Y.P. Bacteria attachment to surfaces—AFM force spectroscopy and physicochemical analyses. J. Colloid Interface Sci. 2011, 364, 213–218. [Google Scholar] [CrossRef]
- Benoit, M.; Gabriel, D.; Gerisch, G.; Gaub, H.E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2000, 2, 313–317. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Grotewold, E.; Vivanco, J.M. Root exudation and rhizosphere biology. Plant. Physiol. 2003, 132, 44–51. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, D.D.; Liu, Y.P.; Li, S.Q.; Shen, Q.R.; Zhang, R.F. Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant. Soil 2014, 374, 689–700. [Google Scholar] [CrossRef]
- Shukla, S.K.; Rao, T.S. Effect of calcium on Staphylococcus aureus biofilm architecture: A confocal laser scanning microscopic study. Colloid Surf. B 2013, 103, 448–454. [Google Scholar] [CrossRef]
- Singh, S.N.; Kumari, B.; Upadhyay, S.K.; Mishra, S.; Kumar, D. Bacterial degradation of pyrene in minimal salt medium mediated by catechol dioxygenases: Enzyme purification and molecular size determination. Bioresour. Technol. 2013, 133, 293–300. [Google Scholar] [CrossRef]
- de Avila, E.D.; Vergani, C.E.; Mollo, F.A.; Jafelicci, M.; Shi, W.Y.; Lux, R. Effect of titanium and zirconia dental implant abutments on a cultivable polymicrobial saliva community. J. Prosthet. Dent. 2017, 118, 481–487. [Google Scholar] [CrossRef] [Green Version]
- Saeki, D.; Nagashima, Y.; Sawada, I.; Matsuyama, H. Effect of hydrophobicity of polymer materials used for water purification membranes on biofilm formation dynamics. Colloid Surf. A 2016, 506, 622–628. [Google Scholar] [CrossRef]
- Pang, C.M.; Hong, P.Y.; Guo, H.L.; Liu, W.T. Biofilm formation characteristics of bacterial isolates retrieved from a reverse osmosis membrane. Environ. Sci. Technol. 2005, 39, 7541–7550. [Google Scholar] [CrossRef]
- Pasmore, M.; Todd, P.; Smith, S.; Baker, D.; Silverstein, J.; Coons, D.; Bowman, C.N. Effects of ultrafiltration membrane surface properties on Pseudomonas aeruginosa biofilm initiation for the purpose of reducing biofouling. J. Membr. Sci. 2001, 194, 15–32. [Google Scholar] [CrossRef]
- dos Reis-Teixeira, F.B.; Alves, V.F.; de Martinis, E.C.P. Growth, viability and architecture of biofilms of Listeria monocytogenes formed on abiotic surfaces. Braz. J. Microbiol. 2017, 48, 587–591. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Ivleva, N.P.; Wagner, M.; Horn, H.; Niessner, R.; Haisch, C. Towards a nondestructive chemical characterization of biofilm matrix by Raman microscopy. Anal. Bioanal. Chem. 2009, 393, 197–206. [Google Scholar] [CrossRef]
- Beech, I.B.; Gubner, R.; Zinkevich, V.; Hanjangsit, L.; Avci, R. Characterisation of conditioning layers formed by exopolymeric substances of Pseudomonas NCIMB 2021 on surfaces of AISI 316 stainless steel. Biofouling 2000, 16, 93–104. [Google Scholar] [CrossRef]
- Wang, H.H.; Ding, S.J.; Wang, G.Y.; Xu, X.L.; Zhou, G.H. In situ characterization and analysis of Salmonella biofilm formation under meat processing environments using a combined microscopic and spectroscopic approach. Int. J. Food Microbiol. 2013, 167, 293–302. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Wang, F.; Zhu, X.S.; Zeng, J.G.; Zhao, Q.G.; Jiang, X. Extracellular polymeric substances govern the development of biofilm and mass transfer of polycyclic aromatic hydrocarbons for improved biodegradation. Bioresour. Technol. 2015, 193, 274–280. [Google Scholar] [CrossRef]
- Mangwani, N.; Kumari, S.; Das, S. Involvement of quorum sensing genes in biofilm development and degradation of polycyclic aromatic hydrocarbons by a marine bacterium Pseudomonas aeruginosa N6P6. Appl. Microbiol. Biotechnol. 2015, 99, 10283–10297. [Google Scholar] [CrossRef]






| Biofilm Parameters | Glass | ‘Liaojing401’ | ‘Koshihikari’ | ‘Zhenzhuhong’ | Polystyrene |
|---|---|---|---|---|---|
| Total biomass (μm3·μm−2) | 0.022 ± 0.005 | 4.400 ± 0.331 | 5.121 ± 0.132 | 6.387 ± 0.470 | 7.519 ± 0.548 |
| Roughness coefficient | 1.975 ± 0.005 | 0.057 ± 0.007 | 0.059 ± 0.013 | 0.056 ± 0.011 | 0.005 ± 0.001 |
| Surface to biovolume Ratio (μm2·μm−3) | 0.113 ± 0.014 | 1.125 ± 0.195 | 1.297 ± 0.088 | 1.495 ± 0.521 | 1.500 ± 0.333 |
| Average diffusion distance (μm) | 0.001 ± 0.000 | 0.923 ± 0.041 | 1.006 ± 0.119 | 1.034 ± 0.000 | 1.265 ± 0.197 |
| Average thickness (μm)) | 12.70 ± 1.71 | 21.50 ± 0.95 | 25.80 ± 2.45 | 30.90 ± 2.54 | 31.70 ± 2.11 |
| Protein Content (μg) | Carbohydrates Content (μg) | |
|---|---|---|
| Glass (10 cm2) | 366.0 ± 4.71 | 122.0 ± 1.19 |
| Polystyrene (10 cm2) | 555.6 ± 3.71 | 149.1 ± 0.92 |
| ‘Liaojing401’ (12 per) | 323.2 ± 3.94 | 258.4 ± 0.79 |
| ‘Koshihikari’ (12 per) | 426.4 ± 5.17 | 272.2 ± 1.28 |
| ‘Zhenzhuhong’ (12 per) | 531.9 ± 6.50 | 309.8 ± 0.97 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Gao, X. Characterization of Biofilm Formed by Phenanthrene-Degrading Bacteria on Rice Root Surfaces for Reduction of PAH Contamination in Rice. Int. J. Environ. Res. Public Health 2019, 16, 2002. https://doi.org/10.3390/ijerph16112002
Zhou Y, Gao X. Characterization of Biofilm Formed by Phenanthrene-Degrading Bacteria on Rice Root Surfaces for Reduction of PAH Contamination in Rice. International Journal of Environmental Research and Public Health. 2019; 16(11):2002. https://doi.org/10.3390/ijerph16112002
Chicago/Turabian StyleZhou, Yuman, and Xiaorong Gao. 2019. "Characterization of Biofilm Formed by Phenanthrene-Degrading Bacteria on Rice Root Surfaces for Reduction of PAH Contamination in Rice" International Journal of Environmental Research and Public Health 16, no. 11: 2002. https://doi.org/10.3390/ijerph16112002
APA StyleZhou, Y., & Gao, X. (2019). Characterization of Biofilm Formed by Phenanthrene-Degrading Bacteria on Rice Root Surfaces for Reduction of PAH Contamination in Rice. International Journal of Environmental Research and Public Health, 16(11), 2002. https://doi.org/10.3390/ijerph16112002