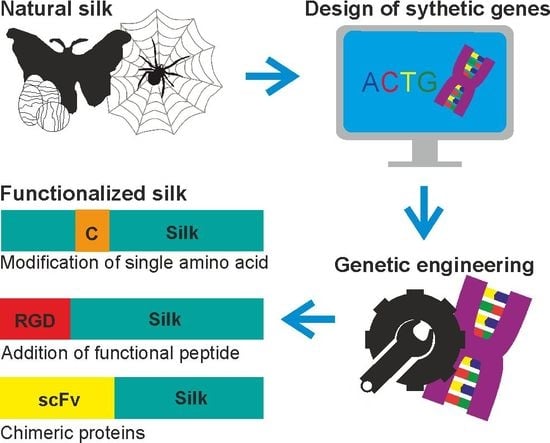
Silk Materials Functionalized via Genetic Engineering for Biomedical Applications
Abstract
:1. Introduction
2. Silk Proteins and Their Recombinant Variants
3. Functionalization of Silk by Changing Its Amino Acid Sequence
4. Functionalization of Silk by Addition of Functional Peptides
4.1. Functionalization of Silk for Cellular Targeting of the Drug Delivery Systems
4.2. Functionalization of Silk for Cell Adhesion
4.3. Functionalization of Silk with Anti-Microbial Properties
4.4. Functionalization of Silk for Binding Inorganic Molecules
5. Functionalization of Silk by Designing Chimeric Proteins
5.1. Chimeric Biopolymers
5.2. Silk Chimeric Proteins for Binding Inorganic Molecules
5.3. Silk Chimeric Proteins for Binding Organic Molecules
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Shao, Z.; Vollrath, F. Materials: Surprising strength of silkworm silk. Nature 2002, 418, 741. [Google Scholar] [CrossRef] [PubMed]
- Altman, G.H.; Diaz, F.; Jakuba, C.; Calabro, T.; Horan, R.L.; Chen, J.; Lu, H.; Richmond, J.; Kaplan, D.L. Silk-based biomaterials. Biomaterials 2003, 24, 401–416. [Google Scholar] [CrossRef]
- Kluge, J.A.; Rabotyagova, O.; Leisk, G.G.; Kaplan, D.L. Spider silks and their applications. Trends Biotechnol. 2008, 26, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Romer, L.; Scheibel, T. The elaborate structure of spider silk: Structure and function of a natural high performance fiber. Prion 2008, 2, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Vendrely, C.; Scheibel, T. Biotechnological production of spider-silk proteins enables new applications. Macromol. Biosci. 2007, 7, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Nileback, L.; Hedin, J.; Widhe, M.; Floderus, L.S.; Krona, A.; Bysell, H.; Hedhammar, M. Self-assembly of recombinant silk as a strategy for chemical-free formation of bioactive coatings: A real-time study. Biomacromolecules 2017, 18, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Schacht, K.; Scheibel, T. Processing of recombinant spider silk proteins into tailor-made materials for biomaterials applications. Curr. Opin. Biotechnol. 2014, 29, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Vepari, C.; Kaplan, D.L. Silk as a biomaterial. Prog. Polym. Sci. 2007, 32, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Kaplan, D.L. Silk-based delivery systems of bioactive molecules. Adv. Drug Deliv. Rev. 2010, 62, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F. Strength and structure of spiders’ silks. J. Biotechnol. 2000, 74, 67–83. [Google Scholar] [CrossRef]
- Heim, M.; Keerl, D.; Scheibel, T. Spider silk: From soluble protein to extraordinary fiber. Angew. Chem. Int. Ed. 2009, 48, 3584–3596. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Porter, D.; Vollrath, F. Morphology and structure of silkworm cocoons. Mater. Sci. Eng. C 2012, 32, 772–778. [Google Scholar] [CrossRef]
- Wray, L.S.; Hu, X.; Gallego, J.; Georgakoudi, I.; Omenetto, F.G.; Schmidt, D.; Kaplan, D.L. Effect of processing on silk-based biomaterials: Reproducibility and biocompatibility. J. Biomed. Mater. Res. B 2011, 99, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Aramwit, P.; Kanokpanont, S.; De-Eknamkul, W.; Srichana, T. Monitoring of inflammatory mediators induced by silk sericin. J. Biosci. Bioeng. 2009, 107, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Acharya, C.; Ghosh, S.K.; Kundu, S.C. Silk fibroin protein from mulberry and non-mulberry silkworms: Cytotoxicity, biocompatibility and kinetics of l929 murine fibroblast adhesion. J. Mater. Sci. Mater. Med. 2008, 19, 2827–2836. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, B.; Taghipour, M.; Rahmani, H.; Sadrjavadi, K.; Fattahi, A. Preparation and characterization of silk fibroin/oligochitosan nanoparticles for sirna delivery. Colloids Surf. B 2015, 136, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Tokareva, O.; Jacobsen, M.; Buehler, M.; Wong, J.; Kaplan, D.L. Structure-function-property-design interplay in biopolymers: Spider silk. Acta Biomater. 2014, 10, 1612–1626. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.V. Spider silk: Ancient ideas for new biomaterials. Chem. Rev. 2006, 106, 3762–3774. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lewis, R.V. Structure of a protein superfiber: Spider dragline silk. Proc. Natl. Acad. Sci. USA 1990, 87, 7120–7124. [Google Scholar] [CrossRef] [PubMed]
- Gaines, W.A.; Sehorn, M.G.; Marcotte, W.R., Jr. Spidroin N-terminal domain promotes a pH-dependent association of silk proteins during self-assembly. J. Biol. Chem. 2010, 285, 40745–40753. [Google Scholar] [CrossRef] [PubMed]
- Ittah, S.; Cohen, S.; Garty, S.; Cohn, D.; Gat, U. An essential role for the C-terminal domain of a dragline spider silk protein in directing fiber formation. Biomacromolecules 2006, 7, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Wang, H.; Wei, K.; Yang, Y.; Zheng, R.-Y.; Kim, I.S.; Zhang, K.-Q. A review of structure construction of silk fibroin biomaterials from single structures to multi-level structures. Int. J. Mol. Sci. 2017, 18, 237. [Google Scholar] [CrossRef] [PubMed]
- Arcidiacono, S.; Mello, C.; Kaplan, D.; Cheley, S.; Bayley, H. Purification and characterization of recombinant spider silk expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 1998, 49, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Fahnestock, S.R.; Irwin, S.L. Synthetic spider dragline silk proteins and their production in Escherichia coli. Appl. Microbiol. Biotechnol. 1997, 47, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Candelas, G.; Candelas, T.; Ortiz, A.; Rodriguez, O. Translational pauses during a spider fibroin synthesis. Biochem. Biophys. Res. Commun. 1983, 116, 1033–1038. [Google Scholar] [CrossRef]
- Lewis, R.V.; Hinman, M.; Kothakota, S.; Fournier, M.J. Expression and purification of a spider silk protein: A new strategy for producing repetitive proteins. Protein Expr. Purif. 1996, 7, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Tokareva, O.; Michalczechen-Lacerda, V.A.; Rech, E.L.; Kaplan, D.L. Recombinant DNA production of spider silk proteins. Appl. Microbiol. Biotechnol. 2013, 6, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Guhrs, K.H.; Grosse, F.; Conrad, U. Production of spider silk proteins in tobacco and potato. Nat. Biotechnol. 2001, 19, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Lazaris, A.; Arcidiacono, S.; Huang, Y.; Zhou, J.F.; Duguay, F.; Chretien, N.; Welsh, E.A.; Soares, J.W.; Karatzas, C.N. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science 2002, 295, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Kuwana, Y.; Sezutsu, H.; Nakajima, K.; Tamada, Y.; Kojima, K. High-toughness silk produced by a transgenic silkworm expressing spider (Araneus ventricosus) dragline silk protein. PLoS ONE 2014, 9, e105325. [Google Scholar] [CrossRef] [PubMed]
- Fahnestock, S.R.; Bedzyk, L.A. Production of synthetic spider dragline silk protein in Pichia pastoris. Appl. Microbiol. Biotechnol. 1997, 47, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Heidebrecht, A.; Scheibel, T. Recombinant production of spider silk proteins. In Advances in Applied Microbiology; Sima, S., Geoffrey, M.G., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 82, pp. 115–153. [Google Scholar]
- Williams, D. Sows’ ears, silk purses and goats’ milk: New production methods and medical applications for silk. Med. Device Technol. 2003, 14, 9–11. [Google Scholar] [PubMed]
- Xia, X.-X.; Qian, Z.-G.; Ki, C.S.; Park, Y.H.; Kaplan, D.L.; Lee, S.Y. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. USA 2010, 107, 14059–14063. [Google Scholar] [CrossRef] [PubMed]
- Dinjaski, N.; Kaplan, D.L. Recombinant protein blends: Silk beyond natural design. Curr. Opin. Biotechnol. 2016, 39, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.D.; Campbell, P.M.; Weisman, S.; Trueman, H.E.; Sriskantha, A.; Wanjura, W.J.; Haritos, V.S. A highly divergent gene cluster in honey bees encodes a novel silk family. Genome Res. 2006, 16, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Sezutsu, H.; Kajiwara, H.; Kojima, K.; Mita, K.; Tamura, T.; Tamada, Y.; Kameda, T. Identification of four major hornet silk genes with a complex of alanine-rich and serine-rich sequences in Vespa simillima xanthoptera Cameron. Biosci. Biotechnol. Biochem. 2007, 71, 2725–2734. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Lua, S.; Du, N.; Liu, X.; Song, J. Identification, recombinant production and structural characterization of four silk proteins from the asiatic honeybee apis cerana. Biomaterials 2008, 29, 2820–2828. [Google Scholar] [CrossRef] [PubMed]
- Weisman, S.; Haritos, V.S.; Church, J.S.; Huson, M.G.; Mudie, S.T.; Rodgers, A.J.; Dumsday, G.J.; Sutherland, T.D. Honeybee silk: Recombinant protein production, assembly and fiber spinning. Biomaterials 2010, 31, 2695–2700. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.D.; Church, J.S.; Hu, X.; Huson, M.G.; Kaplan, D.L.; Weisman, S. Single honeybee silk protein mimics properties of multi-protein silk. PLoS ONE 2011, 6, e16489. [Google Scholar] [CrossRef] [PubMed]
- Krishnaji, S.T.; Bratzel, G.; Kinahan, M.E.; Kluge, J.A.; Staii, C.; Wong, J.Y.; Buehler, M.J.; Kaplan, D.L. Sequence-structure-property relationships of recombinant spider silk proteins: Integration of biopolymer design, processing, and modeling. Adv. Funct. Mater. 2013, 23, 241–253. [Google Scholar] [CrossRef]
- Tokareva, O.S.; Lin, S.; Jacobsen, M.M.; Huang, W.; Rizzo, D.; Li, D.; Simon, M.; Staii, C.; Cebe, P.; Wong, J.Y.; et al. Effect of sequence features on assembly of spider silk block copolymers. J. Struct. Biol. 2014, 186, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Jastrzebska, K.; Felcyn, E.; Kozak, M.; Szybowicz, M.; Buchwald, T.; Pietralik, Z.; Jesionowski, T.; Mackiewicz, A.; Dams-Kozlowska, H. The method of purifying bioengineered spider silk determines the silk sphere properties. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Ryu, S.; Tokareva, O.; Gronau, G.; Jacobsen, M.M.; Huang, W.; Rizzo, D.J.; Li, D.; Staii, C.; Pugno, N.M.; et al. Predictive modelling-based design and experiments for synthesis and spinning of bioinspired silk fibres. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Ebrahimi, D.; Dinjaski, N.; Tarakanova, A.; Buehler, M.J.; Wong, J.Y.; Kaplan, D.L. Synergistic integration of experimental and simulation approaches for the de novo design of silk-based materials. Acc. Chem. Res. 2017, 50, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Szela, S.; Avtges, P.; Valluzzi, R.; Winkler, S.; Wilson, D.; Kirschner, D.; Kaplan, D.L. Reduction-oxidation control of beta-sheet assembly in genetically engineered silk. Biomacromolecules 2000, 1, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Jeannine, M.; Coburn, E.N.; David, L. Kaplan Modulation of vincristine and doxorubicin binding and release from silk films. J. Control. Release 2015, 220, 229–238. [Google Scholar]
- Seib, F.P.; Kaplan, D.L. Doxorubicin-loaded silk films: Drug-silk interactions and in vivo performance in human orthotopic breast cancer. Biomaterials 2012, 33, 8442–8450. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Cebe, P.; Kaplan, D.L. Mechanism of enzymatic degradation of beta-sheet crystals. Biomaterials 2010, 31, 2926–2933. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Lu, C.-L.; Coburn, J.; Kaplan, D.L. Impact of silk biomaterial structure on proteolysis. Acta Biomater. 2015, 11, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Elsner, M.B.; Herold, H.M.; Muller-Herrmann, S.; Bargel, H.; Scheibel, T. Enhanced cellular uptake of engineered spider silk particles. Biomater. Sci. 2015, 3, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Spieß, K.; Wohlrab, S.; Scheibel, T. Structural characterization and functionalization of engineered spider silk films. Soft Matter 2010, 6, 4168–4174. [Google Scholar] [CrossRef]
- Kronqvist, N.; Sarr, M.; Lindqvist, A.; Nordling, K.; Otikovs, M.; Venturi, L.; Pioselli, B.; Purhonen, P.; Landreh, M.; Biverstål, H.; et al. Efficient protein production inspired by how spiders make silk. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Kaplan, D.L. Silk-based gene carriers with cell membrane destabilizing peptides. Biomacromolecules 2010, 11, 3189–3195. [Google Scholar] [CrossRef] [PubMed]
- Kozlowska, A.K.; Florczak, A.; Smialek, M.; Dondajewska, E.; Mackiewicz, A.; Kortylewski, M.; Dams-Kozlowska, H. Functionalized bioengineered spider silk spheres improve nuclease resistance and activity of oligonucleotide therapeutics providing a strategy for cancer treatment. Acta Biomater. 2017, 59, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Florczak, A.; Mackiewicz, A.; Dams-Kozlowska, H. Functionalized spider silk spheres as drug carriers for targeted cancer therapy. Biomacromolecules 2014, 15, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Seib, F.P.; Jones, G.T.; Rnjak-Kovacina, J.; Lin, Y.; Kaplan, D.L. pH-dependent anticancer drug release from silk nanoparticles. Adv. Healthc. Mater. 2013, 2, 1606–1611. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shao, Z.; Chen, X. Paclitaxel-loaded silk fibroin nanospheres. J. Biomed. Mater. Res. A 2012, 100, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Aseh, A.; Ríos, C.N.; Aggarwal, B.B.; Mathur, A.B. Fabrication and characterization of silk fibroin-derived curcumin nanoparticles for cancer therapy. Int. J. Nanomed. 2009, 4, 115–122. [Google Scholar] [CrossRef]
- Qu, J.; Liu, Y.; Yu, Y.; Li, J.; Luo, J.; Li, M. Silk fibroin nanoparticles prepared by electrospray as controlled release carriers of cisplatin. Mater. Sci. Eng. C 2014, 44, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Mochida, A.; Choyke, P.L.; Kobayashi, H. Nanodrug delivery: Is the enhanced permeability and retention effect sufficient for curing cancer? Bioconjug. Chem. 2016, 27, 2225–2238. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Subramanian, B.; Currie, H.A.; Kaplan, D.L. Bioengineered silk protein-based gene delivery systems. Biomaterials 2009, 30, 5775–5784. [Google Scholar] [CrossRef] [PubMed]
- Vannucci, L.; Lai, M.; Chiuppesi, F.; Ceccherini-Nelli, L.; Pistello, M. Viral vectors: A look back and ahead on gene transfer technology. New Microbiol. 2013, 36, 1–22. [Google Scholar] [PubMed]
- Numata, K.; Hamasaki, J.; Subramanian, B.; Kaplan, D.L. Gene delivery mediated by recombinant silk proteins containing cationic and cell binding motifs. J. Control. Release 2010, 146, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Mieszawska-Czajkowska, A.J.; Kvenvold, L.A.; Kaplan, D.L. Silk-based nanocomplexes with tumor-homing peptides for tumor-specific gene delivery. Macromol. Biosci. 2012, 12, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Reagan, M.R.; Goldstein, R.H.; Rosenblatt, M.; Kaplan, D.L. Spider silk-based gene carriers for tumor cell-specific delivery. Bioconjug. Chem. 2011, 22, 1605–1610. [Google Scholar] [CrossRef] [PubMed]
- Watson, K.; Edwards, R.J. HIV-1-trans-activating (Tat) protein: Both a target and a tool in therapeutic approaches. Biochem. Pharmacol. 1999, 58, 1521–1528. [Google Scholar] [CrossRef]
- Rittner, K.; Benavente, A.; Bompard-Sorlet, A.; Heitz, F.; Divita, G.; Brasseur, R.; Jacobs, E. New basic membrane-destabilizing peptides for plasmid-based gene delivery in vitro and in vivo. Mol. Ther. 2002, 5, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Laakkonen, P.; Vuorinen, K. Homing peptides as targeted delivery vehicles. Integr. Biol. 2010, 2, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.A.; Giraudo, E.; Singh, M.; Zhang, L.; Inoue, M.; Porkka, K.; Hanahan, D.; Ruoslahti, E. Progressive vascular changes in a transgenic mouse model of squamous cell carcinoma. Cancer Cell 2003, 4, 383–391. [Google Scholar] [CrossRef]
- Porkka, K.; Laakkonen, P.; Hoffman, J.A.; Bernasconi, M.; Ruoslahti, E. A fragment of the HMGN2 protein homes to the nuclei of tumor cells and tumor endothelial cells in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 7444–7449. [Google Scholar] [CrossRef] [PubMed]
- Florczak, A.; Jastrzebska, K.; Mackiewicz, A.; Dams-Kozlowska, H. Blending two bioengineered spider silks to develop cancer targeting spheres. J. Mater. Chem. B 2017, 5, 3000–3011. [Google Scholar] [CrossRef]
- Witton, C.J.; Reeves, J.R.; Going, J.J.; Cooke, T.G.; Bartlett, J.M. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J. Pathol. 2003, 200, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Bini, E.; Foo, C.W.; Huang, J.; Karageorgiou, V.; Kitchel, B.; Kaplan, D.L. RGD-functionalized bioengineered spider dragline silk biomaterial. Biomacromolecules 2006, 7, 3139–3145. [Google Scholar] [CrossRef] [PubMed]
- Widhe, M.; Johansson, U.; Hillerdahl, C.-O.; Hedhammar, M. Recombinant spider silk with cell binding motifs for specific adherence of cells. Biomaterials 2013, 34, 8223–8234. [Google Scholar] [CrossRef] [PubMed]
- Johansson, U.; Ria, M.; Avall, K.; Dekki Shalaly, N.; Zaitsev, S.V.; Berggren, P.O.; Hedhammar, M. Pancreatic islet survival and engraftment is promoted by culture on functionalized spider silk matrices. PLoS ONE 2015, 10, e0130169. [Google Scholar] [CrossRef] [PubMed]
- Wohlrab, S.; Müller, S.; Schmidt, A.; Neubauer, S.; Kessler, H.; Leal-Egaña, A.; Scheibel, T. Cell adhesion and proliferation on RGD-modified recombinant spider silk proteins. Biomaterials 2012, 33, 6650–6659. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Isozaki, M.; Saotome, T.; Tatematsu, K.-I.; Sezutsu, H.; Kuwabara, N.; Nakazawa, Y. Recombinant silk fibroin incorporated cell-adhesive sequences produced by transgenic silkworm as a possible candidate for use in vascular graft. J. Mater. Chem. B 2014, 2, 7375–7383. [Google Scholar] [CrossRef]
- Widhe, M.; Shalaly, N.D.; Hedhammar, M. A fibronectin mimetic motif improves integrin mediated cell biding to recombinant spider silk matrices. Biomaterials 2016, 74, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Main, A.L.; Harvey, T.S.; Baron, M.; Boyd, J.; Campbell, I.D. The three-dimensional structure of the tenth type III module of fibronectin: An insight into RGD-mediated interactions. Cell 1992, 71, 671–678. [Google Scholar] [CrossRef]
- Gomes, S.C.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Antimicrobial functionalized genetically engineered spider silk. Biomaterials 2011, 32, 4255–4266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, S.; Gallego-Llamas, J.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Biological responses to spider silk-antibiotic fusion protein. J. Tissue Eng. Regen. Med. 2012, 6, 356–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senior, L.; Crump, M.P.; Williams, C.; Booth, P.J.; Mann, S.; Perriman, A.W.; Curnow, P. Structure and function of the silicifying peptide R5. J. Mater. Chem. B 2015, 3, 2607–2614. [Google Scholar] [CrossRef] [Green Version]
- Wong Po Foo, C.; Patwardhan, S.V.; Belton, D.J.; Kitchel, B.; Anastasiades, D.; Huang, J.; Naik, R.R.; Perry, C.C.; Kaplan, D.L. Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 9428–9433. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Huang, W.; Belton, D.J.; Simmons, L.O.; Perry, C.C.; Wang, X.; Kaplan, D.L. Control of silicification by genetically engineered fusion proteins: Silk-silica binding peptides. Acta Biomater. 2015, 15, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Mieszawska, A.J.; Nadkarni, L.D.; Perry, C.C.; Kaplan, D.L. Nanoscale control of silica particle formation via silk-silica fusion proteins for bone regeneration. Chem. Mater. 2010, 22, 5780–5785. [Google Scholar] [CrossRef] [PubMed]
- Dinjaski, N.; Plowright, R.; Zhou, S.; Belton, D.J.; Perry, C.C.; Kaplan, D.L. Osteoinductive recombinant silk fusion proteins for bone regeneration. Acta Biomater. 2017, 49, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Currie, H.A.; Deschaume, O.; Naik, R.R.; Perry, C.C.; Kaplan, D.L. Genetically engineered chimeric silk-silver binding proteins. Adv. Funct. Mater. 2011, 21, 2889–2895. [Google Scholar] [CrossRef] [PubMed]
- Krishnaji, S.T.; Kaplan, D.L. Bioengineered chimeric spider silk-uranium binding proteins. Macromol. Biosci. 2013, 13, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Pardoux, R.; Sauge-Merle, S.; Lemaire, D.; Delangle, P.; Guilloreau, L.; Adriano, J.-M.; Berthomieu, C. Modulating uranium binding affinity in engineered calmodulin EF-hand peptides: Effect of phosphorylation. PLoS ONE 2012, 7, e41922. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, J.A.; Ghandehari, H. Silk-elastinlike protein polymers for matrix-mediated cancer gene therapy. Adv. Drug Deliv. Rev. 2010, 62, 1509–1523. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Tanaka, C.; Yamauchi, K.; Ohgo, K.; Kurokawa, M.; Asakura, T. Silklike materials constructed from sequences of bombyx mori silk fibroin, fibronectin, and elastin. J. Biomed. Mater. Res. A 2008, 84, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-X.; Xu, Q.; Hu, X.; Qin, G.; Kaplan, D.L. Tunable self-assembly of genetically engineered silk-elastin-like protein polymers. Biomacromolecules 2011, 12, 3844–3850. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Huang, Y.; Teng, W.; Cohn, C.M.; Cappello, J.; Wu, X. Complete recombinant silk-elastinlike protein-based tissue scaffold. Biomacromolecules 2010, 11, 3219–3227. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Rollett, A.; Kaplan, D.L. Silk-elastin-like protein biomaterials for the controlled delivery of therapeutics. Expert Opin. Drug Deliv. 2015, 12, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Nagarsekar, A.; Crissman, J.; Crissman, M.; Ferrari, F.; Cappello, J.; Ghandehari, H. Genetic synthesis and characterization of pH- and temperature-sensitive silk-elastinlike protein block copolymers. J. Biomed. Mater. Res. A 2002, 62, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xia, X.; Huang, W.; Lin, Y.; Xu, Q.; Kaplan, D.L. High throughput screening of dynamic silk-elastin-like protein biomaterials. Adv. Funct. Mater. 2014, 24, 4303–4310. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Gustafson, J.A.; MacKay, J.A. Genetically engineered nanocarriers for drug delivery. Int. J. Nanomed. 2014, 9, 1617–1626. [Google Scholar]
- Rozario, T.; DeSimone, D.W. The extracellular matrix in development and morphogenesis: A dynamic view. Dev. Biol. 2010, 341, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Singla, A.; Lee, Y. Biomedical applications of collagen. Int. J. Pharm. 2001, 221, 1–22. [Google Scholar] [CrossRef]
- Ramshaw, J.A.; Peng, Y.Y.; Glattauer, V.; Werkmeister, J.A. Collagens as biomaterials. J. Mater. Sci. Mater. Med. 2009, 20 (Suppl. 1), S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Brodsky, B.; Inouye, M. Dissecting a bacterial collagen domain from streptococcus pyogenes: Sequence and length-dependent variations in triple helix stability and folding. J. Biol. Chem. 2011, 286, 18960–18968. [Google Scholar] [CrossRef] [PubMed]
- An, B.; DesRochers, T.M.; Qin, G.; Xia, X.; Thiagarajan, G.; Brodsky, B.; Kaplan, D.L. The influence of specific binding of collagen-silk chimeras to silk biomaterials on HMSC behavior. Biomaterials 2013, 34, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Wlodarczyk-Biegun, M.K.; Werten, M.W.; de Wolf, F.A.; van den Beucken, J.J.; Leeuwenburgh, S.C.; Kamperman, M.; Cohen Stuart, M.A. Genetically engineered silk-collagen-like copolymer for biomedical applications: Production, characterization and evaluation of cellular response. Acta Biomater. 2014, 10, 3620–3629. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wong, C.; George, A.; Kaplan, D.L. The effect of genetically engineered spider silk-dentin matrix protein 1 chimeric protein on hydroxyapatite nucleation. Biomaterials 2007, 28, 2358–2367. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Spider silk-bone sialoprotein fusion proteins for bone tissue engineering. Soft Matter 2011, 7, 4964–4973. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, M.; Imai, T.; Fujisawa, R.; Tani, H.; Kuboki, Y. Bone sialoprotein (BSP) is a crucial factor for the expression of osteoblastic phenotypes of bone marrow cells cultured on type I collagen matrix. Calcif. Tissue Int. 2000, 66, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.; Gallego-Llamas, J.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. In vivo biological responses to silk proteins functionalized with bone sialoprotein. Macromol. Biosci. 2013, 13, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Jansson, R.; Thatikonda, N.; Lindberg, D.; Rising, A.; Johansson, J.; Nygren, P.A.; Hedhammar, M. Recombinant spider silk genetically functionalized with affinity domains. Biomacromolecules 2014, 15, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Thatikonda, N.; Delfani, P.; Jansson, R.; Petersson, L.; Lindberg, D.; Wingren, C.; Hedhammar, M. Genetic fusion of single-chain variable fragments to partial spider silk improves target detection in micro- and nanoarrays. Biotechnol. J. 2016, 11, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Jansson, R.; Courtin, C.M.; Sandgren, M.; Hedhammar, M. Rational design of spider silk materials genetically fused with an enzyme. Adv. Funct. Mater. 2015, 25, 5343–5352. [Google Scholar] [CrossRef]
- Meirovitch, S.; Shtein, Z.; Ben-Shalom, T.; Lapidot, S.; Tamburu, C.; Hu, X.; Kluge, J.A.; Raviv, U.; Kaplan, D.L.; Shoseyov, O. Spider silk-CBD-cellulose nanocrystal composites: Mechanism of assembly. Int. J. Mol. Sci. 2016, 17, 1573. [Google Scholar] [CrossRef] [PubMed]
- Jastrzebska, K.; Kucharczyk, K.; Florczak, A.; Dondajewska, E.; Mackiewicz, A.; Dams-Kozlowska, H. Silk as an innovative biomaterial for cancer therapy. Rep. Pract. Oncol. Radiother. 2015, 20, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Natural and genetically engineered proteins for tissue engineering. Prog. Polym. Sci. 2012, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, L.; Chen, J.; Wang, L.; Gui, X.; Ran, J.; Xu, G.; Zhao, H.; Zeng, M.; Ji, J.; et al. Silk fibroin biomaterial shows safe and effective wound healing in animal models and a randomized controlled clinical trial. Adv. Healthc. Mater. 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Shtatland, T.; Guettler, D.; Kossodo, M.; Pivovarov, M.; Weissleder, R. Pepbank—A database of peptides based on sequence text mining and public peptide data sources. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Functionalization | Peptide | Bioengineered Silk/Origin | Function of Peptide | Structure | Reference |
|---|---|---|---|---|---|
| Tumor targeting | CGKRK | 6-mer/MaSp1 N. clavipes | Targeting tumor vessels | Complexes pDNA/silk | [66] |
| F3 | 1-mer, 6-mer/MaSp1 N. clavipes | Targeting nucleolin | Complexes pDNA/silk | [65,66] | |
| Lyp1 | 1-mer/MaSp1 N. clavipes | Targeting lymphatic vessels | Complexes pDNA/silk | [65] | |
| H2.1 | MS1/MaSp1 N. clavipes MS2/MaSp2 N. clavipes | Targeting Her2+ receptor | Spheres | [56,72] | |
| H2.2 | MS1/MaSp1 N. clavipes MS2/MaSp2 N. clavipes | Targeting Her2+ receptor | Spheres | [56,72] | |
| Cellular uptake | R8G | eADF4(C16)/ADF4 A. diadematus | Cell penetrating | Spheres | [51] |
| KN | MS2/MaSp2 N. clavipes | Cell penetrating | Spheres | [55] | |
| K15 | 6-mer/MaSp1 N. clavipes | Cell penetrating | Complexes pDNA/silk | [62] | |
| RGD | eADF4(C16)/ADF4 A. diadematus | Targeting integrins | Spheres | [51] | |
| 6-mer/MaSp1 N. clavipes | Targeting integrins | Complexes pDNA/silk | [64] | ||
| ppTG1 | 6-mer/MaSp1 N. clavipes | Cell penetrating | Complexes pDNA/silk | [54] | |
| Tat | eADF4(C16)/ADF4 A. diadematus | Cell penetrating | Spheres | [51] | |
| Nucleic acid binding | K15 | 1-mer, 6-mer/MaSp1 N. clavipes | Binding nucleic acids | Complexes pDNA/silk | [62,64,65,66] |
| KN | MS2/MaSp2 N. clavipes | Binding nucleic acids | Complexes CpG-siRNA/silk, spheres | [55] | |
| Cell binding | IKVAV | 4RepCT/MaSp1 E. australis | Targeting integrins | Fibers, films and foams | [75,76] |
| YIGSR | 4RepCT/MaSp1 E. australis | Targeting integrins | Scaffold | [76] | |
| Light chain/B. mori | Targeting integrins | Films, sponges | [79] | ||
| Heavy chain/B. mori | Targeting integrins | Films, sponges | [79] | ||
| RGD | eADF4(C16)/ADF4 A. diadematus | Targeting integrins | Films | [77] | |
| 4RepCT/MaSp1 E. australis | Targeting integrins | Films | [80] | ||
| 4RepCT/MaSp1 E. australis | Targeting integrins | Fibers, films, foams | [75,76] | ||
| 4RepCT/MaSp1 E. australis | Targeting integrins | Coatings, fibers | [6] | ||
| 15-mer/MaSp1 N. clavipes | Targeting integrins | Fibers, films | [74] | ||
| Heavy chain/B. mori | Targeting integrins | Films, sponges | [79] | ||
| Light chain/B. mori | Targeting integrins | Films, sponges | [79] | ||
| Anti-microbial | Mag | 4RepCT/MaSp1 E. australis | Anti-microbial | Coatings, fibers | [6] |
| HNP-2 | 6-mer/MaSp1 N. clavipes | Anti-microbial | Films | [82] | |
| HNP-4 | 6-mer/MaSp1 N. clavipes | Anti-microbial | Films | [82] | |
| Hepcidin | 6-mer/MaSp1 N. clavipes | Anti-microbial | Films | [82,83] | |
| Inorganic molecules binding | R5 | 15-mer/MaSp1 N. clavipes | Binding silica | Films, fibers, | [85,87] |
| 6-mer/MaSp1 N. clavipes | Binding silica | Soluble, films | [86] | ||
| A1 | 6-mer/MaSp1 N. clavipes | Binding silica | Soluble, films | [86] | |
| A3 | 6-mer/MaSp1 N. clavipes | Binding silica | Soluble, films | [86] | |
| VTK | 15-mer/MaSp1 N. clavipes | Binding hydroxyapatite | Films | [88] | |
| Metal binding | Ag-4 | 6-mer, 15-mer/MaSp1 N. clavipes | Binding silver | Films | [89] |
| Ag-P35 | 6-mer/MaSp1 N. clavipes | Binding silver | Films | [89] | |
| U1 | 6-mer/MaSp1 N. clavipes | Binding uranium | Soluble | [90] | |
| U2 | 6-mer/MaSp1 N. clavipes | Binding uranium | Soluble | [90] |
| Functionalization | Motif/Domain | Bioengineered Silk/Origin | Function of Incorporated Motif/Domain | Structure | Reference |
|---|---|---|---|---|---|
| Chimeric biopolymers | Elastin | (GAGAGS)6/B. mori | Cell binding, drug binding/release, stimuli responsive material | Hydrogels, particles | [92,93,94,95,96,97,98,99] |
| Collagen | (GAGAGS)n/B. mori | Cell binding | Films, scaffolds | [104] | |
| Histidine-rich silk/B. mori | Stimuli responsive material | Hydrogels | [105] | ||
| Inorganic molecules binding | BSP | 6-mer/MaSp1/N. clavipes | Binding hydroxyapatite | Films | [107,109] |
| DMP1 | 15-mer/MaSp1/N. clavipes | Binding hydroxyapatite | Films | [106] | |
| Organic molecules binding | ABD | 4RepCt/MaSp1/E. australis | Binding albumin | Fibers, films | [110] |
| M4 | 4RepCt/MaSp1/E. australis | Binding biotin | Fibers, films | [110] | |
| C2 | 4RepCt/MaSp1/E. australis | Binding IgG | Fibers, films | [110] | |
| Z | 4RepCt/MaSp1/E. australis | Binding IgG | Fibers, films | [110] | |
| scFv | 4RepCt/MaSp1/E. australis | Specific binding of molecules | Fibers | [111] | |
| CBD | 15-mer/MaSp1/N. clavipes | Binding cellulose | Films | [113] | |
| Enzyme | Xylanase | 4RepCt/MaSp1/E. australis | Degradation of polysacharides | Fibers, films, foams | [112] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deptuch, T.; Dams-Kozlowska, H. Silk Materials Functionalized via Genetic Engineering for Biomedical Applications. Materials 2017, 10, 1417. https://doi.org/10.3390/ma10121417
Deptuch T, Dams-Kozlowska H. Silk Materials Functionalized via Genetic Engineering for Biomedical Applications. Materials. 2017; 10(12):1417. https://doi.org/10.3390/ma10121417
Chicago/Turabian StyleDeptuch, Tomasz, and Hanna Dams-Kozlowska. 2017. "Silk Materials Functionalized via Genetic Engineering for Biomedical Applications" Materials 10, no. 12: 1417. https://doi.org/10.3390/ma10121417
APA StyleDeptuch, T., & Dams-Kozlowska, H. (2017). Silk Materials Functionalized via Genetic Engineering for Biomedical Applications. Materials, 10(12), 1417. https://doi.org/10.3390/ma10121417