C60 Bioconjugation with Proteins: Towards a Palette of Carriers for All pH Ranges
Abstract
:1. Introduction
- (i)
- the covalent approach is the most used method to prevent fullerene aggregation. The benefits obtained by functionalization are often offset by reduced photophysical performances [19];
- (ii)
- the noncovalent approach requires the use of supramolecular hosts that are amphipathic molecules able to interact with a single fullerene and to screen it from the aqueous environment. A variety of hosts is capable of interacting with fullerenes. They include surfactants, synthetic polymers, biopolymers, cyclodextrin [20], to name a few. In all cases, they stabilize small clusters of fullerenes [21]. In recent years, also proteins have become used as dispersing agents of fullerenes [9,22,23,24], CNTs [25,26,27,28,29] and graphene [30]. Proteins are naturally amphiphilic. This feature may avoid complicated synthetic procedures or the use of organic solvents. Most proteins are also pH responsive, which is an advantage for some manipulations [26]. Steric hindrance and electrostatic repulsion are the key factors determining the stability of the dispersion of carbon nanomaterials-protein complexes in aqueous solutions [31].
2. Materials and Methods
2.1. C60@Protein Synthesis
2.2. C60@Protein Characterization
2.3. Computational Protocol
3. Results and Discussions
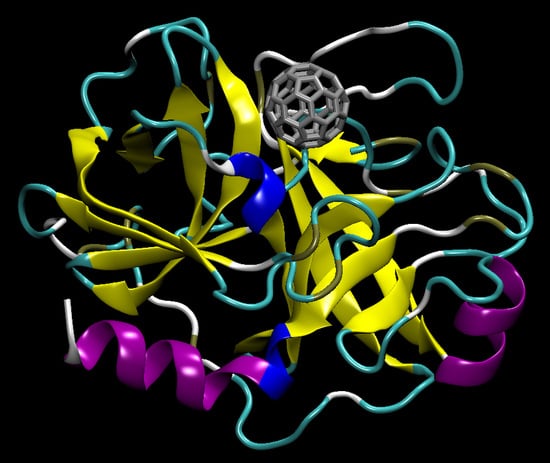
3.1. C60@Pepsin—C60@trypsin, an Atomistic View
- (i)
- (ii)
- Hydrophobic interactions (leucine, isoleucine, methionine, proline, glycine) that are established in water between aliphatic residues and C60 surface [25];
- (iii)
3.2. AFM Analysis of C60@Protein Hybrids
3.3. Stability of the Complex in Aqueous Media
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Nakamura, E.; Isobe, H. Functionalized fullerenes in water. The First 10 years of their chemistry, Biology, and Nanoscience. Acc. Chem. Res. 2003, 36, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, S.; Da Ros, T.; Conde, J.; Sefat, F.; Mozafari, M. Fullerene: Biomedical engineers get to revisit an old friend. Mater. Today 2017, 20, 460–480. [Google Scholar] [CrossRef]
- Castro, E.; Garcia, A.H.; Zavala, G.; Echegoyen, L. Fullerenes in biology and medicine. J. Mater. Chem. B 2017, 5, 6523–6535. [Google Scholar] [CrossRef] [PubMed]
- Dellinger, A.; Zhou, Z.; Connor, J.; Madhankumar, A.; Pamujula, S.; Sayes, C.M.; Kepley, C.L. Application of fullerenes in nanomedicine: An update. Nanomedicine 2013, 8, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Partha, R.; Conyers, J.L. Biomedical applications of functionalized fullerene-based nanomaterials. Int. J. Nanomedicine 2009, 4, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Bosi, S.; Da Ros, T.; Spalluto, G.; Prato, M. Fullerene derivatives: An attractive tool for biological applications. Eur. J. Med. Chem. 2003, 38, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Montellano, A.; Da Ros, T.; Bianco, A.; Prato, M. Fullerene C60 as a multifunctional system for drug and gene delivery. Nanoscale 2011, 3, 4035–4041. [Google Scholar] [CrossRef] [PubMed]
- Mroz, P.; Tegos, G.P.; Gali, H.; Wharton, T.; Sarna, T.; Hamblin, M.R. Fullerenes as photosensitizers in photodynamic therapy. In Medicinal Chemistry and Pharmacological Potential of Fullerenes and Carbon Nanotubes. Carbon Materials: Chemistry and Physics; Springer: Dordrecht, The Netherlands, 2008; pp. 79–106. [Google Scholar]
- Calvaresi, M.; Zerbetto, F. Baiting proteins with C60. ACS Nano 2010, 4, 2283–2299. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-T.; Wang, H.; Guo, L.; Gao, Y.; Liu, Y.; Cao, A. Interaction of fullerenol with lysozyme investigated by experimental and computational approaches. Nanotechnology 2008, 19, 395101. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, P.; Paraskar, A.; Soni, S.; Mashelkar, R.A.; Sengupta, S. Fullerenol−Cytotoxic conjugates for cancer chemotherapy. ACS Nano 2009, 3, 2505–2514. [Google Scholar] [CrossRef] [PubMed]
- Bolskar, R.D. Gadolinium endohedral metallofullerene-based MRI contrast agents. In Medicinal Chemistry and Pharmacological Potential of Fullerenes and Carbon Nanotubes. Carbon Materials: Chemistry and Physics; Springer: Dordrecht, The Netherlands, 2008; pp. 157–180. [Google Scholar]
- Da Ros, T. Twenty years of promises: Fullerene in medicinal chemistry. In Medicinal Chemistry and Pharmacological Potential of Fullerenes and Carbon Nanotubes. Carbon Materials: Chemistry and Physics; Springer: Dordrecht, The Netherlands, 2008; pp. 1–21. [Google Scholar]
- Sharma, S.K.; Chiang, L.Y.; Hamblin, M.R. Photodynamic therapy with fullerenes in vivo: Reality or a dream? Nanomedicine 2011, 6, 1813–1825. [Google Scholar] [CrossRef] [PubMed]
- Mroz, P.; Tegos, G.P.; Gali, H.; Wharton, T.; Sarna, T.; Hamblin, M.R. Photodynamic therapy with fullerenes. Photochem. Photobiol. Sci. 2007, 6, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Guldi, D.M.; Hungerbuehler, H.; Asmus, K.-D. Redox and excitation studies with C60-substituted malonic acid diethyl esters. J. Phys. Chem. 1995, 99, 9380–9385. [Google Scholar] [CrossRef]
- Guldi, D.M.; Prato, M. Excited-state properties of C60 fullerene derivatives. Acc. Chem. Res. 2000, 33, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.L.; Naebe, M.; Barrow, C.J.; Puri, M. Enzyme immobilisation on amino-functionalised multi-walled carbon nanotubes: Structural and biocatalytic characterisation. PLoS ONE 2013, 8, e73642. [Google Scholar] [CrossRef] [PubMed]
- Hamano, T.; Okuda, K.; Mashino, T.; Hirobe, M.; Arakane, K.; Ryu, A.; Mashiko, S.; Nagano, T. Singlet oxygen production from fullerene derivatives: Effect of sequential functionalization of the fullerene core. Chem. Commun. 1997, 21–22. [Google Scholar] [CrossRef]
- Ikeda, A. Water-soluble fullerenes using solubilizing agents, and their applications. J. Incl. Phenom. Macrocycl. Chem. 2013, 77, 49–65. [Google Scholar] [CrossRef]
- Dallavalle, M.; Leonzio, M.; Calvaresi, M.; Zerbetto, F. Explaining fullerene dispersion by using micellar solutions. ChemPhysChem 2014, 15, 2998–3005. [Google Scholar] [CrossRef] [PubMed]
- Vance, S.J.; Desai, V.; Smith, B.O.; Kennedy, M.W.; Cooper, A. Aqueous solubilization of C60 fullerene by natural protein surfactants, latherin and ranaspumin-2. Biophys. Chem. 2016, 214–215, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Arnesano, F.; Bonacchi, S.; Bottoni, A.; Calò, V.; Conte, S.; Falini, G.; Fermani, S.; Losacco, M.; Montalti, M.; et al. C60@Lysozyme: Direct observation by nuclear magnetic resonance of a 1:1 fullerene protein adduct. ACS Nano 2014, 8, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Zerbetto, F. Fullerene sorting proteins. Nanoscale 2011, 3, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Zerbetto, F. The devil and holy water: Protein and carbon nanotube hybrids. Acc. Chem. Res. 2013, 46, 2454–2463. [Google Scholar] [CrossRef] [PubMed]
- Nepal, D.; Geckeler, K.E. pH-Sensitive dispersion and debundling of single-walled carbon nanotubes: Lysozyme as a tool. Small 2006, 2, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Saito, T.; Okazaki, T.; Ohshima, S.; Yumura, M.; Iijima, S. Selectivity of water-soluble proteins in single-walled carbon nanotube dispersions. Chem. Phys. Lett. 2006, 429, 497–502. [Google Scholar] [CrossRef]
- Nepal, D.; Geckeler, K.E. Proteins and Carbon Nanotubes: Close Encounter in Water. Small 2007, 3, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Karajanagi, S.S.; Yang, H.; Asuri, P.; Sellitto, E.; Dordick, J.S.; Kane, R.S. Protein-assisted solubilization of single-walled carbon nanotubes. Langmuir 2006, 22, 1392–1395. [Google Scholar] [CrossRef] [PubMed]
- Joseph, D.; Tyagi, N.; Ghimire, A.; Geckeler, K.E. A direct route towards preparing pH-sensitive graphene nanosheets with anti-cancer activity. RSC Adv. 2014, 4, 4085–4093. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Piella, J.; Bastús, N.G.; Puntes, V. Size-dependent protein–nanoparticle interactions in ctrate-stabilized gold nanoparticles: The emergence of the protein corona. Bioconjugate Chem. 2017, 28, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Boselli, L.; Polo, E.; Castagnola, V.; Dawson, K.A. Regimes of biomolecular ultrasmall nanoparticle interactions. Angew. Chem. Int. Ed. 2017, 56, 4215–4218. [Google Scholar] [CrossRef] [PubMed]
- Bekdemir, A.; Stellacci, F. A centrifugation-based physicochemical characterization method for the interaction between proteins and nanoparticles. Nat. Commun. 2016, 7, 13121. [Google Scholar] [CrossRef] [PubMed]
- Soldà, A.; Cantelli, A.; Di Giosia, M.; Montalti, M.; Zerbetto, F.; Rapino, S.; Calvaresi, M. C60@lysozyme: A new photosensitizing agent for photodynamic therapy. J. Mater. Chem. B 2017, 1757, 525–534. [Google Scholar] [CrossRef]
- Ge, C.; Du, J.; Zhao, L.; Wang, L.; Liu, Y.; Li, D.; Yang, Y.; Zhou, R.; Zhao, Y.; Chai, Z.; et al. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. USA 2011, 108, 16968–16973. [Google Scholar] [CrossRef] [PubMed]
- Walczyk, D.; Bombelli, F.B.; Monopoli, M.P.; Lynch, I.; Dawson, K.A. What the cell “Sees” in bionanoscience. J. Am. Chem. Soc. 2010, 132, 5761–5768. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Greene, D.; Xiao, L.; Qi, R.; Luo, R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2017, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Polak, V.; Shatsky, M.; Halperin, I.; Benyamini, H.; Barzilai, A.; Dror, O.; Haspel, N.; Nussinov, R.; et al. Taking geometry to its edge: Fast unbound rigid (and hinge-bent) docking. Proteins 2003, 52, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Proteins 2007, 69, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Kingsford, C.L.; Chazelle, B.; Singh, M. Solving and analyzing side-chain positioning problems using linear and integer programming. Bioinformatics 2005, 21, 1028–1039. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Vasmatzis, G.; Cornette, J.L.; DeLisi, C. Determination of atomic desolvation energies from the structures of crystallized proteins. J. Mol. Biol. 1997, 267, 707–726. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Darden, T.A.; Cheatham, E.T.; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. AMBER 12, University of California, San Francisco. 2012. Available online: http://ambermd.org/CiteAmber.php (accessed on 27 April 2018).
- Tsui, V.; Case, D.A. Theory and applications of the generalized born solvation model in macromolecular simulations. Biopolymers 2000, 56, 275–291. [Google Scholar] [CrossRef]
- Calvaresi, M.; Bottoni, A.; Zerbetto, F. Thermodynamics of binding between proteins and carbon nanoparticles: The Case of C60@Lysozyme. J. Phys. Chem. C 2015, 119, 28077–28082. [Google Scholar] [CrossRef]
- Calvaresi, M.; Furini, S.; Domene, C.; Bottoni, A.; Zerbetto, F. Blocking the passage: C60 geometrically clogs K+ Channels. ACS Nano 2015, 9, 4827–4834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trozzi, F.; Marforio, T.D.; Bottoni, A.; Zerbetto, F.; Calvaresi, M. Engineering the fullerene-protein interface by computational design: The sum is more than its Parts. Isr. J. Chem. 2017, 57, 547–552. [Google Scholar] [CrossRef]
- Jonsson, M. Isoelectric spectra of native and base denatured crystallized swine pepsin. Acta Chem. Scand. 1972, 26, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.A. Trypsinogens and trypsins of various species. Methods Enzymol. 1970, 19, 41–63. [Google Scholar] [CrossRef]
- Chen, Z.; Westerhoff, P.; Herckes, P. Quantification of C60 fullerene concentrations in water. Environ. Toxicol. Chem. 2008, 27, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.H.; DeCamp, D.L.; Sijbesma, R.P.; Srdanov, G.; Wudl, F.; Kenyon, G.L. Inhibition of the HIV-1 protease by fullerene derivatives: Model building studies and experimental verification. J. Am. Chem. Soc. 1993, 115, 6506–6509. [Google Scholar] [CrossRef]
- Sijbesma, R.; Srdanov, G.; Wudl, F.; Castoro, J.A.; Wilkins, C.; Friedman, S.H.; DeCamp, D.L.; Kenyon, G.L. Synthesis of a fullerene derivative for the inhibition of HIV enzymes. J. Am. Chem. Soc. 1993, 115, 6510–6512. [Google Scholar] [CrossRef]
- Marcorin, G.L.; Da Ros, T.; Castellano, S.; Stefancich, G.; Bonin, I.; Miertus, S.; Prato, M. Design and synthesis of novel [60]Fullerene Derivatives as Potential HIV Aspartic Protease Inhibitors. Org. Lett. 2000, 2, 3955–3958. [Google Scholar] [CrossRef] [PubMed]
- Schuster, D.I.; Wilson, S.R.; Schinazi, R.F. Anti-human immunodeficiency virus activity and cytotoxicity of derivatized buckminsterfullerenes. Bioorganic Med. Chem. Lett. 1996, 6, 1253–1256. [Google Scholar] [CrossRef]
- Tokuyama, H.; Yamago, S.; Nakamura, E.; Shiraki, T.; Sugiura, Y. Photoinduced biochemical activity of fullerene carboxylic acid. J. Am. Chem. Soc. 1993, 115, 7918–7919. [Google Scholar] [CrossRef]
- Iwashita, K.; Shiraki, K.; Ishii, R.; Tanaka, T.; Hirano, A. Liquid chromatographic analysis of the interaction between amino acids and aromatic surfaces using single-wall carbon nanotubes. Langmuir 2015, 31, 8923–8929. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Hoefinger, S.; Zerbetto, F. Probing the structure of lysozyme-carbon-nanotube hybrids with molecular dynamics. Chem. A Eur. J. 2012, 18, 4308–4313. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Tanaka, T.; Kataura, H.; Kameda, T. Arginine side chains as a dispersant for individual single-wall carbon nanotubes. Chem. Eur. J. 2014, 20, 4922–4930. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Kameda, T.; Wada, M.; Tanaka, T.; Kataura, H. Carbon nanotubes facilitate oxidation of cysteine residues of proteins. J. Phys. Chem. Lett. 2017, 8, 5216–5221. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Kameda, T.; Sakuraba, S.; Wada, M.; Tanaka, T.; Kataura, H. Disulfide bond formation of thiols by using carbon nanotubes. Nanoscale 2017, 9, 5389–5393. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, S.; Alargova, R.G.; Tsujii, K. Stable dispersions of fullerenes, C60 and C70, in water. preparation and characterization. Langmuir 2001, 17, 6013–6017. [Google Scholar] [CrossRef]
- Tang, Y.J.; Ashcroft, J.M.; Chen, D.; Min, G.; Kim, C.-H.; Murkhejee, B.; Larabell, C.; Keasling, J.D.; Chen, F.F. Charge-associated effects of fullerene derivatives on microbial structural integrity and central metabolism. Nano Lett. 2007, 7, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, D.G.; Efremova, L.V.; Vasilchenko, A.S.; Saidakova, E.V.; Sizova, E.A.; Troshin, P.A.; Zhilenkov, A.V.; Khakina, E.A. A zeta potential value determines the aggregate’s size of penta-substituted [60]fullerene derivatives in aqueous suspension whereas positive charge is required for toxicity against bacterial cells. J. Nanobiotechnol. 2015, 13, 50. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, D.G.; Davydova, O.K.; Yankina, Z.Z.; Vasilchenko, A.S.; Miroshnikov, S.A.; Kornev, A.B.; Ivanchikhina, A.V.; Troshin, P.A. The Activity of [60]Fullerene derivatives bearing amine and carboxylic solubilizing Groups against Escherichia coli: A Comparative Study. J. Nanomater. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Tegos, G.P.; Demidova, T.N.; Arcila-Lopez, D.; Lee, H.; Wharton, T.; Gali, H.; Hamblin, M.R. Cationic fullerenes are effective and selective antimicrobial photosensitizers. Chem. Biol. 2005, 12, 1127–1135. [Google Scholar] [CrossRef] [PubMed]






| C60@protein Complex | Top 10 Residues Interacting with C60 | ||||
|---|---|---|---|---|---|
| C60@Pepsin-Binding pocket 1 | Phe 111 = −5.7 | Leu 112 = −3.1 | Thr 218 = −3.0 | Ser 219 = −2.9 | Thr 12 = −2.8 |
| Glu 13 = −2.8 | Phe 117 = −2.6 | Ile 30 = −2.5 | Tyr 75 = −2.5 | Thr 77 = −2.2 | |
| C60@Pepsin-Binding pocket 2 | Val 291 = −4.9 | Thr 74 = −4.3 | Pro 292 = −3.7 | Tyr 75 = −3.4 | Gly 76 = −2.7 |
| Met 289 = −1.4 | Thr 293 = −1.3 | Tyr 189 = −1.2 | Asp 290 = −1.0 | Leu 298 = −0.6 | |
| C60@Trypsin | His 57 = −4.9 | Phe 41 = −4.2 | Gln 192 = −3.5 | Cys 58 = −3.4 | Cys 42 = −2.7 |
| Gly 193 = −1.8 | Ser 195 = −1.7 | Asp 194 = −0.8 | Tyr 151 = −0.6 | Leu 99 = −0.4 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Giosia, M.; Valle, F.; Cantelli, A.; Bottoni, A.; Zerbetto, F.; Calvaresi, M. C60 Bioconjugation with Proteins: Towards a Palette of Carriers for All pH Ranges. Materials 2018, 11, 691. https://doi.org/10.3390/ma11050691
Di Giosia M, Valle F, Cantelli A, Bottoni A, Zerbetto F, Calvaresi M. C60 Bioconjugation with Proteins: Towards a Palette of Carriers for All pH Ranges. Materials. 2018; 11(5):691. https://doi.org/10.3390/ma11050691
Chicago/Turabian StyleDi Giosia, Matteo, Francesco Valle, Andrea Cantelli, Andrea Bottoni, Francesco Zerbetto, and Matteo Calvaresi. 2018. "C60 Bioconjugation with Proteins: Towards a Palette of Carriers for All pH Ranges" Materials 11, no. 5: 691. https://doi.org/10.3390/ma11050691
APA StyleDi Giosia, M., Valle, F., Cantelli, A., Bottoni, A., Zerbetto, F., & Calvaresi, M. (2018). C60 Bioconjugation with Proteins: Towards a Palette of Carriers for All pH Ranges. Materials, 11(5), 691. https://doi.org/10.3390/ma11050691