Graphene Quantum Dots as Nanozymes for Electrochemical Sensing of Yersinia enterocolitica in Milk and Human Serum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
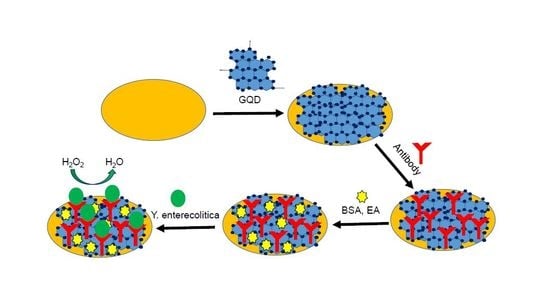
2.2. Fabrication, Cleaning and Coating of Working Electrodes
2.3. Determination of Optimal GQDs Concentration for Bioassays
2.4. Antibody Immobilization and Characterization
2.5. Development of the Bioassay for Milk Samples
2.6. Development of the Bioassay for Human Serum Samples
3. Results and Discussions
3.1. Determination of Optimal GQDs Concentration for Bioassays
3.2. Label Free Direct Assay for Y. enterocolitica Determination in Milk and Human Blood
3.3. Cross-Reactivity Studies for Y. enterocolitica
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rosner, B.M.; Werber, D.; Höhle, M.; Stark, K. Clinical aspects and self- reported symptoms of sequelae Yersinia enterocolitica infections in a population-based study, Germany 2009–2010. BMC Infect. Dis. 2013, 13, 1471–2334. [Google Scholar] [CrossRef] [PubMed]
- EFSA and ECDC. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar]
- Tacket, C.O.; Narain, J.P.; Sattin, R.; Lofgren, J.P.; Konigsberg, C., Jr.; Rendtorff, R.C.; Rausa, A.; Davis, B.R.; Cohen, M.L. A multistate outbreak of infections caused by Yersinia enterocolitica transmitted by pasteurized milk. JAMA 1984, 251, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Love, U.J.; Bradshaw, J.G.; Peeler, J.T. Thermal Inactivation of Yenterocolitica in Milk. Appl. Environ. Microbiol. 1982, 44, 517–519. [Google Scholar]
- Sdliemarın, D.A. Association of Yersinia enterocolitica with the manufacture of cheese and occurrence in pasteurized milk. Appl. Environ. Microbiol. 1978, 36, 274–277. [Google Scholar] [Green Version]
- Hughes, D. Repeated isolation of Yersinia enterocolitica from pasteurized milk in a holding vat at a dairy factory. J. Appl. Bacteriol. 1980, 48, 383–385. [Google Scholar] [CrossRef]
- Tebbs, R.S.; Wong, L.Y.; Brzoska, P.; Petrauskene, O.V. Molecular technologies for Salmonella detection. In Salmonella Distribution, Adaptation, Control Measures and Molecular Technologies; IntechOpen: London, UK, 2012; pp. 481–504. [Google Scholar] [CrossRef]
- Hochel, I.; Skvor, J. Characterization of rabbit antibodies for immunochemical detection of Yersinia enterocolitica. Folia Microbiol. 2007, 52, 511–518. [Google Scholar] [CrossRef]
- Luciani, M.; Schirone, M.; Portanti, O.; Visciano, P.; Armillotta, G.; Tofalo, R.; Suzzi, G.; Sonsini, L.; Di Febo, T. Development of a rapid method for the detection of Yersinia enterocolitica serotype O:8 from food. Food Microbiol. 2018, 73, 85–92. [Google Scholar] [CrossRef]
- Balakrishna, K.; Radhika, M.; Murali, H.S.; Batra, H.V.; Bawa, A.S. Specific identification of pathogenic Yersinia enterocolitica by monoclonal antibodies generated against recombinant attachment invasion locus protein. World J. Microbiol. Biot. 2012, 28, 533–539. [Google Scholar] [CrossRef]
- Stachelska, M.A. Identification of pathogenicity of Yersinia enterocolitica in pig tonsils using the real-time PCR. Pol. J. Microbiol. 2018, 67, 219–222. [Google Scholar] [CrossRef]
- Oh, B.K.; Lee, W.; Chun, B.S.; Bae, Y.M.; Lee, W.H.; Choi, J.W. Surface plasmon resonance immunosensor for the detection of Yersinia enterocolitica. Coll. Surf. A. 2005, 257–258, 369–374. [Google Scholar] [CrossRef]
- Bae, Y.M.; Oh, B.K.; Lee, W.; Lee, W.H.; Choi, J.H. Immunosensorfor detection of Yersinia enterocolitica Based on imaging elipsometry. Anal. Chem. 2004, 76, 1799–1803. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhao, Y.; Bi, Y.; Liu, H.; Guo, Z.; Song, Y.; Zhai, J.; Huang, H.; Yang, R. Direct detection of Yersinia pestis from the infected animal specimens by a fiber optic biosensor. Sens. Actuators B Chem. 2006, 123, 204–210. [Google Scholar] [CrossRef]
- Sun, W.; Qin, P.; Gao, H.; Li, G.; Jiao, K. Electrochemical DNA biosensor based on chitosan/nano-V2O5/MWCNTs composite film modified carbon ionic liquid electrode and its application to the LAMP product of Yersinia enterocolitica gene sequence. Biosens. Bioelectron. 2010, 25, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Brandon, R.; Cate, M.; Peng, X.; Stony, R.; Johnson, M. Detection of pathogens using luminescent CdSe/ZnS dendron nanocrystals and a porous membrane immunofilter. Anal. Chem. 2007, 79, 8796–8802. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of new label-free techniques for biosensors: A review. Crit. Rev. Biotechnol. 2016, 36, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Guo, J.; Bao, X.; Chen, T.; Weng, W.; Chen, S.; Yang, M. Rapid and sensitive detection of bacteria response to antibiotics using nanoporous membrane and graphene quantum dot (GQDs)-based electrochemical biosensors. Materials 2017, 10, 603. [Google Scholar] [CrossRef] [PubMed]
- Hanjun, S.; Wu, L.; Wei, W.; Qu, X. Recent advances in graphene quantum dots for sensing. Mater. Today. 2013, 16, 433–442. [Google Scholar]
- Xiang, Q.; Huang, J.; Huang, H.; Mao, W.; Ye, Z. A label-free electrochemical platform for the highly sensitive detection of hepatitis B virus DNA using graphene quantum dots. RSC Adv. 2018, 8, 1820–1825. [Google Scholar] [CrossRef] [Green Version]
- Xiaoming, L.; Muchen, R.; Jizhong, S.; Zihan, S.; Haibo, Z. Carbon and graphene quantum dots for optoelectronic and energy devices: A review. Adv. Funct. Mater. 2015, 25, 4929–4947. [Google Scholar]
- Zhang, Y.; Wu, C.; Zhou, X.; Wu, X.; Yang, Y.; Wu, H.; Guo, S.; Zhang, J. Garphene quantum dots/gold electrode and its application in living cell H2O2 detection. Nanoscale 2013, 5, 1816–1819. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Q.; Liu, Y.; Chui, J.; Liu, H.; Wang, P.; Li, Y.; Chen, L.; Zhao, Z.; Dong, Y. A novel label-free electrochemical immunosensor based on functionalized nitrogen-doped graphene quantum dots for carcinoembryonic antigen detection. Biosens. Bioelectron. 2017, 90, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Savas, S.; Ersoy, A.; Gulmez, Y.; Kılıc, S.; Levent, B.; Altıntas, Z. Nanoparticle enhanced antibody and DNA biosensors for sensitive detection of Salmonella. Materials 2018, 11, 1541. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z.; Uludag, Y.; Gurbuz, Y.; Tothill, I. Development of surface chemistry for surface plasmon resonance based sensors for the detection of proteins and DNA molecules. Anal. Chim. Acta 2012, 712, 138–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bang, D.; Lee, T.; Park, J.; Lee, G.; Haam, S.; Park, J. Enhancement of capturing efficacy for circulating tumor cells by centrifugation. BioChip J. 2018, 12, 38–45. [Google Scholar] [CrossRef]
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef]
- Komarova, E.; Reber, K.; Aldissi, M.; Bogomolova, A. New multispecific array as a tool for electrochemical impedance spectroscopy-based biosensing. Biosens. Bioelectron. 2010, 25, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Masdor, N.A.; Altintas, Z.; Tothill, I.E. Sensitive detection of Campylobacter jejuni using nanoparticles enhanced QCM sensor. Biosens. Bioelectron. 2016, 78, 328–336. [Google Scholar] [CrossRef]
- Masdor, N.A.; Altintas, Z.; Tothill, I.E. Surface plasmon resonance immunosensor for the detection of Campylobacter jejuni. Chemosensors 2017, 5, 16. [Google Scholar] [CrossRef]
- Magliulo, M.; Simoni, P.; Guardigli, M.; Michelini, E.; Luciani, M.; Lelli, R.; Roda, A. A Rapid multiplexed chemiluminescent immunoassay for the detection of Escherichia coli 0157:H7, Yersinia enterocolitica, Salmonella typhimurium and Listeria monocytogenes pathogen bacteria. J. Agric. Food Chem. 2007, 55, 4933–4939. [Google Scholar] [CrossRef]
- Bonardi, S.; Bruini, I.; D’Incau, M.; Van Damme, I.; Carniel, E.; Bremont, S.; Cavallini, P.; Tagliabue, S.; Brindani, F. Detection, seroprevalence and antimicrobial resistance of Yersinia enterocolitica and Yersinia pseudotuberculosis in pig tonsils in Nothern Italy. Int J. Food Microbiol. 2016, 235, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Bhaduri, S. Comparison of multiplex PCR, PCR-ELISA and fluorogenic 5’ muclease PCR assays for detection of plasmid-bearing virulent Yersinia enterocolitica in swine feces. Mol. Cell. Probes 2002, 16, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, S.; Wacheck, S.; Koenig, M.; Stolle, A.; Stephan, R. Prevalence of pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis in wild boars in Switzerland. Int. J. Food Microbiol. 2009, 135, 199–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddique, N.; Sharma, D.; Al-Khaldi, S.F. Detection of Yersinia enterocolitica in alfalfa, mung bean, cilantro, and mamey sapote (Pouteria sapota) food matrices using DNA microarray chip hybridization. Curr. Microbiol. 2009, 59, 233–239. [Google Scholar] [CrossRef] [PubMed]









| Method | Sample | Ligand | Investigation Range | Detection Limit | Ref |
|---|---|---|---|---|---|
| ELISA | Dairy product | Antibody | 5‒1000 cfu g−1 | 10 cfu g−1 | [9] |
| Chemiluminescent ELISA | Human fecal/ bovine meat | Antibody | 102‒108 cfu mL−1 | 104‒105 cfu mL−1 | [31] |
| ELISA | Pig diaphragm muscle samples | Antibody | 2‒4.75 log10 cfu g−1 | 3.56 log10 cfu g−1 | [32] |
| Multiplex PCR | Pig feces | Ail gene | P/N | P/N | [33] |
| Real-time PCR and ELISA | Wild boar tonsils/feaces | Ail gene | P/N | P/N | [34] |
| SPR sensor | Buffer | Protein G | 102‒107 cfu mL−1 | 102 cfu mL−1 | [12] |
| Imaging ellipsometry immunosensor | Buffer | Antibody | 103‒107 cfu mL−1 | 103 cfu mL−1 | [13] |
| DNA microarray chip hybridization | Various food matrices | DNA | 103‒106 cfu g−1 | 104 cfu g-1 * | [35] |
| GQD sensor | Milk | Antibody | 1‒6.23 × 108 cfu mL−1 | 5 cfu mL−1 | CW |
| GQD sensor | Human serum | Antibody | 1‒6.23 × 108 cfu mL−1 | 30 cfu mL−1 | CW |
| Bacteria Tested (107 cfu mL−1) | Response (Current, nA) | GQD Immunosensor % Relative Activity |
|---|---|---|
| Yersinia enterocolitica | 1473.0 | 100.0 ± 2.0 |
| Yersinia pestis | 93.2 | 6.0 ± 2.0 |
| Salmonella enteritidis | 27.4 | 1.9 ± 0.2 |
| Escherichia coli | 9.2 | 0.6 ± 0.1 |
| Bacillus anthracis | 1.0 | 0.0 ± 0.0 |
| Mix (− Y. pestis) * | 1441.0 | 97.8 ± 1.5 |
| Mix (+ Y. pestis) ** | 1392.0 | 94.5 ± 2.2 |
| Bacteria Tested (107 cfu mL−1) | Response (Current, nA) | GQD Immunosensor % Relative Activity |
|---|---|---|
| Yersinia enterocolitica | 12.0 | 100.0 ± 2.0 |
| Yersinia pestis | 2.7 | 22.8 ± 2.0 |
| Salmonella enteritidis | 1.1 | 9.3 ± 0.3 |
| Escherichia coli | 1.0 | 8.3 ± 0.2 |
| Bacillus anthracis | 0.5 | 4.5 ± 0.1 |
| Mix (− Y. pestis) * | 10.5 | 87.5 ± 1.5 |
| Mix (+ Y. pestis) ** | 9.1 | 75.8 ± 2.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savas, S.; Altintas, Z. Graphene Quantum Dots as Nanozymes for Electrochemical Sensing of Yersinia enterocolitica in Milk and Human Serum. Materials 2019, 12, 2189. https://doi.org/10.3390/ma12132189
Savas S, Altintas Z. Graphene Quantum Dots as Nanozymes for Electrochemical Sensing of Yersinia enterocolitica in Milk and Human Serum. Materials. 2019; 12(13):2189. https://doi.org/10.3390/ma12132189
Chicago/Turabian StyleSavas, Sumeyra, and Zeynep Altintas. 2019. "Graphene Quantum Dots as Nanozymes for Electrochemical Sensing of Yersinia enterocolitica in Milk and Human Serum" Materials 12, no. 13: 2189. https://doi.org/10.3390/ma12132189
APA StyleSavas, S., & Altintas, Z. (2019). Graphene Quantum Dots as Nanozymes for Electrochemical Sensing of Yersinia enterocolitica in Milk and Human Serum. Materials, 12(13), 2189. https://doi.org/10.3390/ma12132189