Highly Selective Fluorescence Sensor Based on Graphene Quantum Dots for Sulfamethoxazole Determination
Abstract
:1. Introduction
2. Experimental Details
2.1. Materials
2.2. Instruments and Measurements
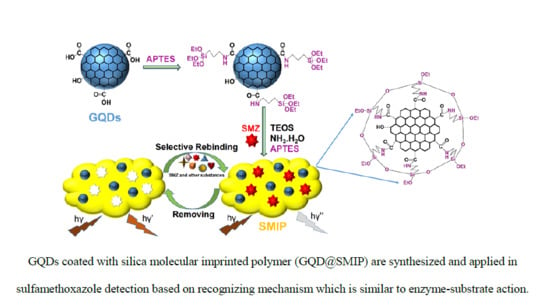
2.3. Synthesis of GQDs and APTES-Modified GQDs
2.4. Synthesis of GQDs@SMIP and silica non-molecularly imprinted polymer (GQDs@SNIP)
2.5. SMZ detection
2.6. Selectivity
3. Results and Discussion
3.1. Characterization of GQDs@SMIP
3.2. SMZ Sensing
3.3. Selectivity
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mookherjee, S.; Shoen, C.; Cynamon, M. In Vitro Activity of JPC 2067 Alone and in Combination with Sulfamethoxazole against Nocardia Species. Antimicrob. Agents Chemother. 2012, 56, 1133–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.Z.; Wang, Y.C.; Huang, W.H.; Yu, L.; Yang, Y.F.; Liu, H.; Yang, W.M. Computer-aided design and synthesis of CdTe@SiO2 core-shell molecularly imprinted polymers as a fluorescent sensor for the selective determination of sulfamethoxazole in milk and lake water. J. Sep. Sci. 2017, 40, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Dirany, A.; Sires, I.; Oturan, N.; Ozcan, A.; Oturan, M.A. Electrochemical Treatment of the Antibiotic Sulfachloropyridazine: Kinetics, Reaction Pathways, and Toxicity Evolution. Environ. Sci. Technol. 2012, 46, 4074–4082. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.H.; Stemig, A.M.; Wammer, K.H.; Strathmann, T.J. Oxidation of Antibiotics during Water Treatment with Potassium Permanganate: Reaction Pathways and Deactivation. Environ. Sci. Technol. 2011, 45, 3635–3642. [Google Scholar] [CrossRef]
- Jansomboon, W.; Boontanon, S.K.; Boontanon, N.; Polprasert, C. Determination and health risk assessment of enrofloxacin, flumequine and sulfamethoxazole in imported Pangasius catfish products in Thailand. J. Environ. Sci. Health Part B Pestic. Contam. Agric. Wastes 2018, 53, 108–115. [Google Scholar] [CrossRef]
- Khalaf, H.; Haidari, P.; Dikran, S.; Mohammed, P. Spectrophotometric Determination of Sulfamethoxazole in Pure and Pharmaceutical Preparations Based on Condensation Reaction Method. J. Babylon Univ. Pure Appl. Sci. 2017, 25, 515–524. [Google Scholar]
- Ait Lahcen, A.; Amine, A. Electrochemical determination of sulfamethoxazole using different kinds of electrodes. In Proceedings of the XXV Congresso Nazionale della Socetà Italiana, Arcavacata di Rendie, Italy, 7–12 September 2014. [Google Scholar]
- Wan, J.; Jin, C.; Liu, B.; Zonglian, S.; Gao, M.; Wang, Z. Electrochemical oxidation of sulfamethoxazole using Ti/SnO2-Sb/Co-PbO2 electrode through ANN-PSO. J. Serb. Chem. Soc. 2019, 84, 25. [Google Scholar] [CrossRef]
- Jank, L.; Martins, M.T.; Arsand, J.B.; Ferrao, M.F.; Hoff, R.B.; Barreto, F.; Pizzolato, T.M. An LC-ESI-MS/MS method for residues of fluoroquinolones, sulfonamides, tetracyclines and trimethoprim in feedingstuffs: Validation and surveillance. Food Addit. Contam. Part A Chem. 2018, 35, 1975–1989. [Google Scholar] [CrossRef]
- Amini, H.; Ahmadiani, A. Rapid and simultaneous determination of sulfamethoxazole and trimethoprim in human plasma by high-performance liquid chromatography. J. Pharm. Biomed. Anal. 2007, 43, 1146–1150. [Google Scholar] [CrossRef]
- Avgerinos, A.; Athanasiou, G.; Malamataris, S. Rapid simultaneous determination of trimethoprim, sulfamethoxazole and acetylsulfamethoxazole in human plasma and urine by high-performance liquid-chromatography. J. Pharm. Biomed. Anal. 1991, 9, 507–510. [Google Scholar] [CrossRef]
- Mogolodi Dimpe, K.; Mpupa, A.; Nomngongo, P.N. Microwave assisted solid phase extraction for separation preconcentration sulfamethoxazole in wastewater using tyre based activated carbon as solid phase material prior to spectrophotometric determination. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 188, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Issac, S.; Kumar, K.G. Voltammetric determination of sulfamethoxazole at a multiwalled carbon nanotube modified glassy carbon sensor and its application studies. Drug Test. Anal. 2009, 1, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, H.J.; Kim, H.M.; Kim, S.H.; Lee, J.W.; Kim, S.K.; Cho, B.R. Metal ion sensing novel calix 4 crown fluoroionophore with a two-photon absorption property. J. Org. Chem. 2006, 71, 8016–8022. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.C.; Wu, J.S.; Kim, H.J.; Kwon, P.S.; Kim, J.W.; Bartsch, R.A.; Lee, J.Y.; Kim, J.S. Rationally designed fluorescence ‘turn-on’ sensor for Cu2+. Chem. Commun. 2011, 47, 3165–3167. [Google Scholar] [CrossRef]
- Lin, L.X.; Zhang, S.W. Creating high yield water soluble luminescent graphene quantum dots via exfoliating and disintegrating carbon nanotubes and graphite flakes. Chem. Commun. 2012, 48, 10177–10179. [Google Scholar] [CrossRef]
- Liu, X.T.; Na, W.D.; Liu, H.; Sue, X.G. Fluorescence turn-off-on probe based on polypyrrole/graphene quantum composites for selective and sensitive detection of paracetamol and ascorbic acid. Biosens. Bioelectron. 2017, 98, 222–226. [Google Scholar] [CrossRef]
- Mehrzad-Samarin, M.; Faridbod, F.; Dezfuli, A.S.; Ganjali, M.R. A novel metronidazole fluorescent nanosensor based on graphene quantum dots embedded silica molecularly imprinted polymer. Biosens. Bioelectron. 2017, 92, 618–623. [Google Scholar] [CrossRef]
- Amjadi, M.; Jalili, R. Molecularly imprinted polymer-capped nitrogen-doped graphene quantum dots as a novel chemiluminescence sensor for selective and sensitive determination of doxorubicin. RSC Adv. 2016, 6, 86736–86743. [Google Scholar] [CrossRef]
- Kazemifard, N.; Ensafi, A.A.; Rezaei, B. Green synthesized carbon dots embedded in silica molecularly imprinted polymers, characterization and application as a rapid and selective fluorimetric sensor for determination of thiabendazole in juices. Food Chem. 2020, 310, 9. [Google Scholar] [CrossRef]
- Liu, H.C.; Ding, J.; Zhang, K.; Ding, L. Fabrication of carbon dots@restricted access molecularly imprinted polymers for selective detection of metronidazole in serum. Talanta 2020, 209, 10. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wu, Y.; Xu, G.; Gao, Y.; Yang, J.; Liu, L.; Zhou, P.; Hu, Q. Molecularly imprinted polymer based on CdTe@SiO2 quantum dots as a fluorescent sensor for the recognition of norepinephrine. Analyst 2014, 139, 5785–5792. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, X.; Liu, J.; Wu, M.; Li, M.; Zang, X. One-pot synthesis of a fluorescent molecularly imprinted nanosensor for highly selective detection of sulfapyridine in water. Anal. Methods 2018, 10, 884–890. [Google Scholar] [CrossRef]
- Huamin, Q.; Lulu, F.; Li, X.; Li, L.; Min, S.; Chuannan, L. Determination sulfamethoxazole based chemiluminescence and chitosan/graphene oxide-molecularly imprinted polymers. Carbohydr. Polym. 2013, 92, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, G.; Xiao, B.; Yan, J. Molecularly imprinted electrochemical sensor for the determination of sulfamethoxazole. J. New Mat. Electrochem. Syst. 2018, 21, 77–80. [Google Scholar] [CrossRef]
- Khanfar, M.F.; Abu-Nameh, E.S.M.; Saket, M.M.; Al Khateeb, L.T.; Al Ahmad, A.; Asaad, Z.; Salem, Z.; Alnuman, N. Detection of Hydrochlorothiazide, Sulfamethoxazole, and Trimethoprim at Metal Oxide Modified Glassy Carbon Electrodes. Int. J. Electrochem. Sci. 2020, 15, 1771–1787. [Google Scholar] [CrossRef]
- Liu, X.; Liu, J.; Huang, Y.; Zhao, R.; Liu, G.; Chen, Y. Determination of methotrexate in human serum by high-performance liquid chromatography combined with pseudo template molecularly imprinted polymer. J. Chromatogr. A 2009, 1216, 7533–7538. [Google Scholar] [CrossRef]
- Balasubramanian, P.; Settu, R.; Chen, S.M.; Chen, T.W. Voltammetric sensing of sulfamethoxazole using a glassy carbon electrode modified with a graphitic carbon nitride and zinc oxide nanocomposite. Microchim. Acta 2018, 185, 9. [Google Scholar] [CrossRef]
- Hou, J.; Li, H.Y.; Wang, L.; Zhang, P.; Zhou, T.Y.; Ding, H.; Ding, L. Rapid microwave-assisted synthesis of molecularly imprinted polymers on carbon quantum dots for fluorescent sensing of tetracycline in milk. Talanta 2016, 146, 34–40. [Google Scholar] [CrossRef]
- Chao, M.R.; Hu, C.W.; Chen, J.L. Comparative syntheses of tetracycline-imprinted polymeric silicate and acrylate on CdTe quantum dots as fluorescent sensors. Biosens. Bioelectron. 2014, 61, 471–477. [Google Scholar] [CrossRef]
- Zhou, Y.; Qu, Z.B.; Zeng, Y.; Zhou, T.; Shi, G. A novel composite of graphene quantum dots and molecularly imprinted polymer for fluorescent detection of paranitrophenol. Biosens. Bioelectron. 2014, 52, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.B.; Qin, X.Y.; Liu, S.; Chang, G.H.; Zhang, Y.W.; Luo, Y.L.; Asiri, A.M.; Al-Youbi, A.O.; Sun, X.P. Economical, Green Synthesis of Fluorescent Carbon Nanoparticles and Their Use as Probes for Sensitive and Selective Detection of Mercury(II) Ions. Anal. Chem. 2012, 84, 5351–5357. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Sun, Z.C.; Zheng, M.; Li, J.; Zhang, Y.Q.; Zhang, G.Q.; Zhao, H.F.; Liu, X.Y.; Xie, Z.G. Three Colors Emission from S,N Co-doped Graphene Quantum Dots for Visible Light H-2 Production and Bioimaging. Adv. Opt. Mater. 2015, 3, 360–367. [Google Scholar] [CrossRef]
- Xu, Q.; Li, B.F.; Ye, Y.C.; Cai, W.; Li, W.J.; Yang, C.Y.; Chen, Y.S.; Xu, M.; Li, N.; Zheng, X.S.; et al. Synthesis, mechanical investigation, and application of nitrogen and phosphorus co-doped carbon dots with a high photoluminescent quantum yield. Nano Res. 2018, 11, 3691–3701. [Google Scholar] [CrossRef]
- Wu, S.-H.; Mou, C.-Y.; Lin, H.-P. Synthesis of Mesoporous Silica Nanoparticles. Chem. Soc. Rev. 2013, 42, 3862–3875. [Google Scholar] [CrossRef]
- Robert, W.; Zuhl, Z.A. Solution Chemistry Impact on Silica Polymerization by Inhibitors; Mineral Scales in Biological and Industrial Systems; Amjad, Z., Ed.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2013. [Google Scholar]
- Lan, Y.K.; Chen, T.C.; Tsai, H.J.; Wu, H.C.; Lin, J.H.; Lin, I.K.; Lee, J.F.; Chen, C.S. Adsorption Behavior and Mechanism of Antibiotic Sulfamethoxazole on Carboxylic-Functionalized Carbon Nanofibers-Encapsulated Ni Magnetic Nanoparticles. Langmuir 2016, 32, 9530–9539. [Google Scholar] [CrossRef]
- Zhang, H.M.; Gui, Y.Q.; Cao, Y.; Wang, M.; Liu, B.Z. Molecularly Imprinted Sensor based on o-phenylenediamine for Electrochemical Detection of Sulfamethoxazole. Int. J. Electrochem. Sci. 2019, 14, 11630–11640. [Google Scholar] [CrossRef]
- Turco, A.; Corvaglia, S.; Mazzotta, E.; Pompa, P.P.; Malitesta, C. Preparation and characterization of molecularly imprinted mussel inspired film as antifouling and selective layer for electrochemical detection of sulfamethoxazole. Sens. Actuators B Chem. 2018, 255, 3374–3383. [Google Scholar] [CrossRef]
- Lamaoui, A.; Lahcen, A.A.; García-Guzmán, J.J.; Palacios-Santander, J.M.; Cubillana-Aguilera, L.; Amine, A. Study of solvent effect on the synthesis of magnetic molecularly imprinted polymers based on ultrasound probe: Application for sulfonamide detection. Ultrason. Sonochem. 2019, 58, 104670. [Google Scholar] [CrossRef]
- Yari, A.; Shams, A. Silver-filled MWCNT nanocomposite as a sensing element for voltammetric determination of sulfamethoxazole. Anal. Chim. Acta 2018, 1039, 51–58. [Google Scholar] [CrossRef]








| Method | Linear Range | LOD | Reference |
|---|---|---|---|
| Electrochemical | 0.2–1.4 µM | 0.05 µM | [39] |
| Electrochemical | 0.8–170 µM | 0.8 µM | [40] |
| Electrochemical | 2.4–23.6 µM | 0.81 µM | [26] |
| Electrochemical | 10–100 µM | 0.144 µM | [27] |
| Ultrasound | 0.78–19.74 µM | 0.23 µM | [41] |
| Voltammetry | 0.05–70 µM | 0.01 µM | [42] |
| Our method | 1–100 µM | 1 µM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, T.H.; Lee, H.J.; Kim, J.H.; Park, S.J. Highly Selective Fluorescence Sensor Based on Graphene Quantum Dots for Sulfamethoxazole Determination. Materials 2020, 13, 2521. https://doi.org/10.3390/ma13112521
Le TH, Lee HJ, Kim JH, Park SJ. Highly Selective Fluorescence Sensor Based on Graphene Quantum Dots for Sulfamethoxazole Determination. Materials. 2020; 13(11):2521. https://doi.org/10.3390/ma13112521
Chicago/Turabian StyleLe, Thi Hoa, Hyun Jong Lee, Ji Hyeon Kim, and Sang Joon Park. 2020. "Highly Selective Fluorescence Sensor Based on Graphene Quantum Dots for Sulfamethoxazole Determination" Materials 13, no. 11: 2521. https://doi.org/10.3390/ma13112521
APA StyleLe, T. H., Lee, H. J., Kim, J. H., & Park, S. J. (2020). Highly Selective Fluorescence Sensor Based on Graphene Quantum Dots for Sulfamethoxazole Determination. Materials, 13(11), 2521. https://doi.org/10.3390/ma13112521