Multivariate Discriminant Analysis of Single Seed Near Infrared Spectra for Sorting Dead-Filled and Viable Seeds of Three Pine Species: Does One Model Fit All Species?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Measurement of NIR Spectra
2.3. Model Development and Validation
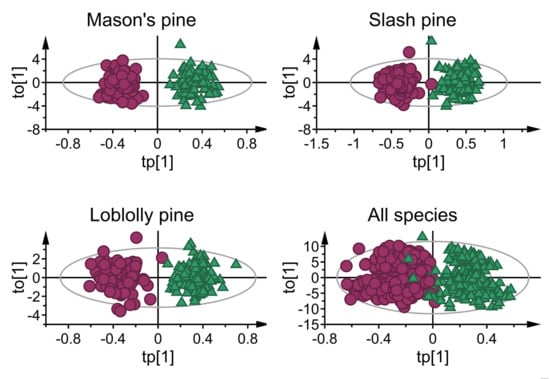
3. Results
3.1. Mean Absorbance Values and Model Overview
3.2. Classification Performance of Fitted Models
3.3. Absorption Bands Relevant for Discriminating Viable and Dead-Filled Pine Seeds
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Payn, T.; Carnus, J.-M.; Freer-Smith, P.; Kimberley, M.; Kollert, W.; Liu, S.; Orazio, C.; Rodriguez, L.; Silva, L.N.; Wingfield, M.J. Changes in planted forests and future global implications. For. Ecol. Manag. 2015, 352, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Del Lungo, A.; Ball, J.; Carle, J. Global Planted Forests Thematic Study: Results and Analysis. Planted Forests and Tree Working Paper 38, FAO, Rome. 2006. Available online: http://www.fao.org/forestry/12139-03441d093f070ea7d7c4e3ec3f306507.pdf (accessed on 20 March 2019).
- Winsa, H.; Sahlén, K. Effects of Seed Invigoration and Microsite Preparation on Seedling Emergence and Establishment After Direct Sowing of Pinus sylvestris L. at Different Dates. Scand. J. For. Res. 2001, 16, 422–428. [Google Scholar] [CrossRef]
- Landis, T.D. The Target Plant Concept―a history and brief overview. In National Proc: Forest and Conservation Nursery Assoc―2010; Riley, L.E., Haase, D.L., Pinto, J.R., Eds.; Tech cords.; Proc RMRS-P-65; US Dept Agric Forest Serv Rocky Mtn Res Sta: Ft Collins, CO, USA, 2011; pp. 61–66. [Google Scholar]
- Dumroese, R.K.; Landis, T.D.; Pinto, J.R.; Haase, D.L.; Wilkinson, K.W.; Davis, A.S. Meeting forest restoration challenges: Using the target plant concept. Reforesta 2016, 1, 37–52. [Google Scholar] [CrossRef]
- Slobodník, B.; Gutternberger, H. Ovule, megaspores and female gametophyte formation in Larix decidua Mill. (Pinaceae). Acta Biologica Cracov. 2000, 42, 93–100. [Google Scholar]
- Gutterman, Y. Maternal effects on seeds during development. In Seeds: The Ecology of Regeneration in Plant Communities, 2nd ed.; Fenner, M., Ed.; CABI Publishing: Wallingford, UK, 2000; pp. 59–84. [Google Scholar]
- Simak, M. A method for the removal of filled-dead seeds from a sample of Pinus contorta. Seed Sci. Technol. 1984, 12, 767–775. [Google Scholar]
- Downie, B.; Wang, B.S.P. Upgrading germinability and vigor of jack pine, logdepole pine, and white spruce by the IDS technique. Can. J. For. Res. 1992, 22, 1124–1131. [Google Scholar] [CrossRef]
- Sivakumar, V.; Anandalakshmi, R.; Warrier, R.R.; Singh, B.G.; Tigabu, M.; Odén, P.C. Petroleum flotation technique upgrades the germinability of Casuarina equisetifolia seed lots. New For. 2007, 34, 281–291. [Google Scholar] [CrossRef]
- Agelet, L.E.; Hurburgh, C.R. Limitations and current applications of Near Infrared Spectroscopy for single seed analysis. Talanta 2014, 121, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Workman, J.; Weyer, L. Practical Guide and Spectral Atlas for Interpretive Near- Infrared Spectroscopy, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2012; pp. 1–303. [Google Scholar]
- Burns, D.A.; Ciurczak, E.W. Handbook of Near-Infrared Analysis, 2nd ed.; Marcel Dekker Inc.: New York, NY, USA, 2001; pp. 1–803. [Google Scholar]
- Tigabu, M.; Odén, P.C. Multivariate classification of sound and insect-infested seeds of a tropical multipurpose tree, Cordia africana, with near infrared reflectance spectroscopy. J. Near Infrared Spectrosc. 2002, 10, 45–51. [Google Scholar] [CrossRef]
- Tigabu, M.; Odén, P.C. Near infrared spectroscopy-based method for separation of sound and insect-damaged seeds of Albizia schimperiana, a multipurpose legume. Seed Sci. Technol. 2003, 31, 317–328. [Google Scholar] [CrossRef]
- Tigabu, M.; Odén, P.C. Simultaneous detection of filled, empty and insect-infested seeds of three Larix species with single seed near infrared transmittance spectroscopy. New For. 2004, 27, 39–53. [Google Scholar] [CrossRef]
- Tigabu, M.; Odén, P.C.; Shen, T.Y. Application of near infrared spectroscopy for the detection of internal insect infestation in Picea abies seed lots. Can. J. For. Res. 2004, 34, 76–84. [Google Scholar] [CrossRef]
- Tigabu, M.; Fjellström, J.; Odén, P.C.; Teketay, D. Germination of Juniperus procera seeds in response to stratification and smoke treatments, and detection of insect-damaged seeds with VIS + NIR spectroscopy. New For. 2007, 33, 155–169. [Google Scholar] [CrossRef]
- Tigabu, M.; Odén, P.C. Classification of viable and empty seeds of Pinus patula Schiede & Deppe with near-infrared spectroscopy and multivariate analysis. New For. 2003, 25, 163–176. [Google Scholar]
- Daneshvar, A.; Tigabu, M.; Karimidoost, A.; Odén, P.C. Single seed near infrared spectroscopy discriminates viable and non-viable seeds of Juniperus polycarpos. Silva Fenn. 2015, 49, 14. [Google Scholar] [CrossRef]
- Farhadi, M.; Tigabu, M.; Odén, P.C. Near Infrared Spectroscopy as non-destructive method for sorting viable, petrified and empty seeds of Larix sibirica. Silva Fenn. 2015, 49, 12. [Google Scholar] [CrossRef]
- Lestander, T.A.; Odén, P.C. Separation of viable and non-viable filled Scots pine seeds by differentiating between drying rates using single seed near infrared transmittance spectroscopy. Seed Sci. Technol. 2002, 30, 383–392. [Google Scholar]
- Trygg, J.; Wold, S. O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integral OSC filter. J. Chemom. 2003, 17, 53–64. [Google Scholar] [CrossRef]
- Baskin, C.C.; Baskin, J.M. Seeds: Ecology, Biogeography and Evolution of Dormancy and Germination, 2nd ed.; Academic Press: San Diego, CA, USA, 2001; pp. 1–666. [Google Scholar]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Rinnan, Å.; Van Den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC-Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (VIP) for OPLS models and its applicability in multivariate time series analysis. Chemom. Intell. Lab. Syst. 2015, 146, 297–304. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi- and Megavariate data analysis: Basic Principles and Applications, 2nd ed.; Umetrics Academy: Umeå, Sweden, 2006; pp. 63–101. [Google Scholar]
- Næs, T.; Isaksson, T.; Fearn, T.; Davies, T. A User Friendly Guide to Multivariate Calibration and Classification; NIR Publications: Chichester, UK, 2002; pp. 1–344. [Google Scholar]
- Tigabu, M.; Farhadi, M.; Stener, L.-G.; Odén, P.C. Visible + Near Infrared Spectroscopy as taxonomic tool for identifying birch species. Silva Fenn. 2018, 52, 13. [Google Scholar] [CrossRef]
- Pinto, R.C.; Trygg, J.; Gottfries, J. Advantages of orthogonal inspection in chemometrics. J. Chemom. 2012, 26, 231–235. [Google Scholar] [CrossRef]
- Shenk, J.S.; Workman, J.J.; Westerhaus, M.O. Application of NIR spectroscopy to agricultural products. In Handbook of Near-Infrared Spectroscopy, 2nd ed.; Burns, D.A., Ciurczak, E.W., Eds.; Marcel Dekker Inc.: New York, NY, USA, 2001; pp. 419–474. [Google Scholar]





| Species | Viable Seeds | Dead-Filled Seeds |
|---|---|---|
| Mason’s pine | 13.9 | 8.0 |
| Slash pine | 13.0 | 6.2 |
| Loblolly pine | 8.1 | 6.1 |
| Species | Model Statistics | ||||
|---|---|---|---|---|---|
| A | R2Xp | R2Xo | R2Y | Q2cv | |
| Mason’s pine | 1 + 9 | 0.026 | 0.974 | 0.942 | 0.923 |
| Loblolly pine | 1 + 5 | 0.024 | 0.976 | 0.901 | 0.864 |
| Slash pine | 1 +7 | 0.021 | 0.979 | 0.909 | 0.879 |
| All species | 1 + 12 | 0.003 | 0.997 | 0.804 | 0.796 |
| All species-truncated spectra | 1 + 10 | 0.02 | 0.980 | 0.811 | 0.797 |
| Species | Class | Sn | Sp | CA | ER |
|---|---|---|---|---|---|
| Mason’s pine | Viable | 1.0 | 1.0 | 1.0 | 0.0 |
| Dead-filled | 1.0 | 1.0 | 1.0 | 0.0 | |
| Loblolly pine | Viable | 1.0 | 1.0 | 1.0 | 0.0 |
| Dead-filled | 1.0 | 1.0 | 1.0 | 0.0 | |
| Slash pine | Viable | 1.0 | 1.0 | 1.0 | 0.0 |
| Dead-filled | 1.0 | 1.0 | 1.0 | 0.0 | |
| All species | Viable | 0.98 | 0.99 | 0.99 | 0.01 |
| Dead-filled | 0.99 | 0.98 | 0.99 | 0.01 | |
| All species: truncated spectra | Viable | 0.99 | 0.99 | 0.99 | 0.01 |
| Dead-filled | 0.98 | 0.98 | 0.99 | 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tigabu, M.; Daneshvar, A.; Jingjing, R.; Wu, P.; Ma, X.; Odén, P.C. Multivariate Discriminant Analysis of Single Seed Near Infrared Spectra for Sorting Dead-Filled and Viable Seeds of Three Pine Species: Does One Model Fit All Species? Forests 2019, 10, 469. https://doi.org/10.3390/f10060469
Tigabu M, Daneshvar A, Jingjing R, Wu P, Ma X, Odén PC. Multivariate Discriminant Analysis of Single Seed Near Infrared Spectra for Sorting Dead-Filled and Viable Seeds of Three Pine Species: Does One Model Fit All Species? Forests. 2019; 10(6):469. https://doi.org/10.3390/f10060469
Chicago/Turabian StyleTigabu, Mulualem, Abolfazl Daneshvar, Ren Jingjing, Pengfei Wu, Xiangqing Ma, and Per Christer Odén. 2019. "Multivariate Discriminant Analysis of Single Seed Near Infrared Spectra for Sorting Dead-Filled and Viable Seeds of Three Pine Species: Does One Model Fit All Species?" Forests 10, no. 6: 469. https://doi.org/10.3390/f10060469
APA StyleTigabu, M., Daneshvar, A., Jingjing, R., Wu, P., Ma, X., & Odén, P. C. (2019). Multivariate Discriminant Analysis of Single Seed Near Infrared Spectra for Sorting Dead-Filled and Viable Seeds of Three Pine Species: Does One Model Fit All Species? Forests, 10(6), 469. https://doi.org/10.3390/f10060469