Development and Characterization of Double-Antibody Sandwich ELISA for Detection of Zika Virus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of the NS1 Protein
2.2. Ethics Statement
2.3. Production of Monoclonal and Polyclonal Antibodies Against the ZIKV-NS1 Protein
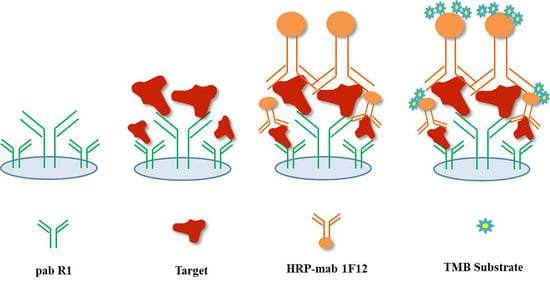
2.4. Detection of Recombinant ZIKV-NS1 Protein by the DAS-ELISA
2.5. Titer and Binding Affinity of mAb 1F12 and pAb R1 Based on the ELISA
2.6. Western Blot Assay
2.7. Indirect Immunofluorescent Assay
2.8. Establishment of the DAS-ELISA Based on pAb R1 and mAb 1F12 Probe
2.9. Specificity and Sensitivity of the DAS-ELISA
2.10. Application of the DAS-ELISA for the Detection of ZIKV Infection in Cells and Tree Shrews
3. Results
3.1. Expression and Purification of the ZIKV-NS1 Protein
3.2. Evaluation of the Rabbit Antiserum
3.3. Characterization of the Ascites Against NS1 Protein
3.4. Establishment of the DAS-ELISA
3.5. Optimization Operations
3.6. The Specificity and Sensitivity of ZIKV Detection by DAS-ELISA
3.7. Application of the DAS-ELISA for ZIKV Detection in the Supernatants, Cell Lysates of Cells, and the Sera of Tree Shrews
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Dick, G.W.A.; Kitchen, S.F.; Haddow, A.J. Zika Virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J.; Honein, M.A.; Petersen, L.R. Zika Virus and Birth Defects—Reviewing the Evidence for Causality. N. Engl. J. Med. 2016, 374, 1981–1987. [Google Scholar] [CrossRef] [PubMed]
- Coyne, C.B.; Lazear, H.M. Zika virus- reigniting the TORCH. Nat. Rev. Microbiol. 2016, 14, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Brasil, P.; Pereira, J.P., Jr.; Moreira, M.E.; Nogueira, R.M.R.; Damasceno, L.; Wakimoto, M.; Rabello, R.S.; Valderramos, S.G.; Halai, U.A.; Salles, T.S. Zika Virus Infection in Pregnant Women in Rio de Janeiro. N. Engl. J. Med. 2016, 375, 2321–2334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caolormeau, V.M.; Blake, A.; Mons, S.; Lastère, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet 2016, 387, 1531–1539. [Google Scholar] [CrossRef]
- Heymann, D.L.; Hodgson, A.; Sall, A.A.; Freedman, D.O.; Staples, J.E.; Althabe, F.; Baruah, K.; Mahmud, G.; Kandun, N.; Vasconcelos, P.F.C. Zika virus and microcephaly: Why is this situation a PHEIC. Lancet 2016, 387, 719–721. [Google Scholar] [CrossRef]
- Fauci, A.S.; Morens, D.M. Zika Virus in the Americas—Yet Another Arbovirus Threat. N. Engl. J. Med. 2016, 374, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Maurer-Stroh, S.; Mak, T.M.; Ng, Y.K.; Phuah, S.P.; Huber, R.G.; Marzinek, J.K.; Holdbrook, D.A.; Lee, R.T.; Cui, L.; Lin, R.T. South-east Asian Zika virus strain linked to cluster of cases in Singapore, August 2016. Eurosurveillance 2016, 21, 3407. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.A.; Mier-Y-Teran-Romero, L.; Reefhuis, J.; Gilboa, S.M.; Hills, S.L. Zika and the Risk of Microcephaly. N. Engl. J. Med. 2016, 375, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, W.; Wong, G.; Bi, Y.; Yan, J.; Yi, S.; Chen, E.; Hao, Y.; Lou, X.; Mao, H. Highly diversified Zika viruses imported to China, 2016. Protein Cell 2016, 7, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Haddow, A.D.; Schuh, A.J.; Yasuda, C.Y.; Kasper, M.R.; Heang, V.; Huy, R.; Guzman, H.; Tesh, R.B.; Weaver, S.C. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis. 2012, 6, e1477. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.K. Zika virus: Epidemiology, current phobia and preparedness for upcoming mass gatherings, with examples from World Olympics and Pilgrimage. Pak. J. Med. Sci. 2016, 32, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Bhavani, K.G.; Krishna, K.B.M.; Srinivasu, N.; Ramachandran, D.; Raman, N.V.V.S.S.; Babu, B.H. Determination of genotoxic impurity in atazanavir sulphate drug substance by LC-MS. J. Pharm. Biomed. Anal. 2017, 132, 156–158. [Google Scholar]
- Nicolini, A.M.; Mccracken, K.E.; Yoon, J.Y. Future developments in biosensors for field-ready Zika virus diagnostics. J. Biol. Eng. 2017, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Gubler, D.J. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, J.T. Update: Interim Guidance for Prevention of Sexual Transmission of Zika Virus—United States, July 2016. Morb. Mortal. Wkly. Rep. 2016, 65, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Diamond, M.S.; Gale, M., Jr. West Nile virus infection and immunity. Nat. Rev. Microbiol. 2013, 11, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Klema, V.; Padmanabhan, R.; Choi, K. Flaviviral Replication Complex: Coordination between RNA Synthesis and 5′-RNA Capping. Viruses 2015, 7, 4640–4656. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Young, P.R.; Hilditch, P.A.; Bletchly, C.; Halloran, W. An Antigen Capture Enzyme-Linked Immunosorbent Assay Reveals High Levels of the Dengue Virus Protein NS1 in the Sera of Infected Patients. J. Clin. Microbiol. 2000, 38, 1053–1057. [Google Scholar] [PubMed]
- Cecchetto, J.; Fernandes, F.C.B.; Lopes, R.; Bueno, P.R. The capacitive sensing of NS1 Flavivirus biomarker. Biosens. Bioelectron. 2017, 87, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Parkash, O.; Shueb, R.H. Diagnosis of Dengue Infection Using Conventional and Biosensor Based Techniques. Viruses 2015, 7, 5410–5427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.J.; Furuya, A.; Zou, J.; Xie, X.; Ii, A.P.D.; Kramer, L.D.; Shi, P.Y. A Multiplex Microsphere Immunoassay for Zika Virus Diagnosis. Ebiomedicine 2017, 16, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F. Specificity, cross-reactivity and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Roche, C.; Nhan, T.X.; Robin, E.; Teissier, A.; Caolormeau, V.M. Detection of Zika virus in saliva. J. Clin. Virol. 2015, 68, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Bingham, A.M. Comparison of Test Results for Zika Virus RNA in Urine, Serum, and Saliva Specimens from Persons with Travel-Associated Zika Virus Disease—Florida, 2016. Morb. Mortal. Wkly. Rep. 2016, 65, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Sun, T.; Xia, X.; Wei, Q.; Song, Y.; Han, Q.; Chen, Q.; Hu, J.; Zhang, J. Optimized Expression, Purification of Herpes B Virus gD Protein in Escherichia coli, and Production of Its Monoclonal Antibodies. Jundishapur J. Microbiol. 2016, 9, e32183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, C.; Dai, L.; Zhang, L.; Xu, K.; Song, Y.; Xia, X.; Han, Q.; Chen, Q.; Zhang, J. Efficient Capture and Detection of Zika Virion by Polyclonal Antibody Against Prokaryotic Recombinant Envelope Protein. Jundishapur J. Microbiol. 2018, 11, e68858. [Google Scholar] [CrossRef]
- Musso, D.; Nilles, E.J.; Cao-Lormeau, V.M. Rapid spread of emerging Zika virus in the Pacific area. Clin. Microbiol. Infect. 2014, 20, O595–O596. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Zeng, H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem. 2017, 89, 12743–12748. [Google Scholar] [CrossRef] [PubMed]
- Langerak, T.; Yang, H.; Baptista, M.; Doornekamp, L.; Kerkman, T.; Codrington, J.; Roosblad, J.; Vreden, S.G.; De Bruin, E.; Mogling, R.; et al. Zika Virus Infection and Guillain-Barre Syndrome in Three Patients from Suriname. Front. Neurol. 2016, 7, 233. [Google Scholar] [CrossRef] [PubMed]
- Gourinat, A.C.; O’Connor, O.; Calvez, E.; Goarant, C.; Dupontrouzeyrol, M. Detection of Zika Virus in Urine. Emerg. Infect. Dis. 2015, 21, 84–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Name | Sequence | Length (bp) |
|---|---|---|
| ZIKV-ASF | GGTCAGCGTCCTCTCTAATAAACG | 24 |
| ZIKV-ASR | GCACCCTAGTGTCCACTTTTTCC | 23 |
| ZIKV-Probe | FAM-AGCCATGACCGACACCACACCGT-BQ1 | 23 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Du, X.; Chen, C.; Chen, Z.; Zhang, L.; Han, Q.; Xia, X.; Song, Y.; Zhang, J. Development and Characterization of Double-Antibody Sandwich ELISA for Detection of Zika Virus Infection. Viruses 2018, 10, 634. https://doi.org/10.3390/v10110634
Zhang L, Du X, Chen C, Chen Z, Zhang L, Han Q, Xia X, Song Y, Zhang J. Development and Characterization of Double-Antibody Sandwich ELISA for Detection of Zika Virus Infection. Viruses. 2018; 10(11):634. https://doi.org/10.3390/v10110634
Chicago/Turabian StyleZhang, Liding, Xuewei Du, Congjie Chen, Zhixin Chen, Li Zhang, Qinqin Han, Xueshan Xia, Yuzhu Song, and Jinyang Zhang. 2018. "Development and Characterization of Double-Antibody Sandwich ELISA for Detection of Zika Virus Infection" Viruses 10, no. 11: 634. https://doi.org/10.3390/v10110634
APA StyleZhang, L., Du, X., Chen, C., Chen, Z., Zhang, L., Han, Q., Xia, X., Song, Y., & Zhang, J. (2018). Development and Characterization of Double-Antibody Sandwich ELISA for Detection of Zika Virus Infection. Viruses, 10(11), 634. https://doi.org/10.3390/v10110634