Highlighting of a LAGLIDADG and a Zing Finger Motifs Located in the pUL56 Sequence Crucial for HCMV Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Conserved Patterns and Secondary Structure Prediction
2.2. Cells and Bacterial Strains
2.3. BAC Mutagenesis
2.4. Reconstitution of Viruses Harboring the Mutations
2.5. Plaque Assays and Growth Curve Analysis
2.6. Viral Immediate Early and Late Proteins Expression
3. Results
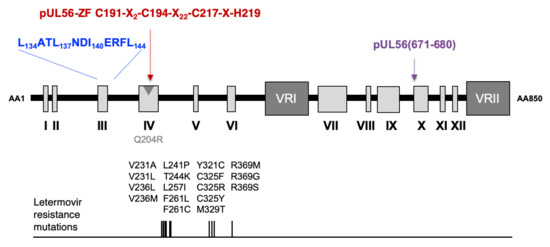
3.1. Identification of a LATLNDIERFL Pattern into pUL56, Characteristic of the LAGLIDADG Homing Endonuclease
3.2. Several Amino Acids of pUL56 Putative 134LATLNDIERFL144 Pattern are Essential for Viral Replication
3.3. The Putative Zinc Finger Pattern of pUL56 is Required for Viral Replication
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alain, S.; Hantz, S.; Scieux, C.; Karras, A.; Mazeron, M.C.; Szelag, J.C.; Imbert, B.M.; Fillet, A.M.; Gouarin, S.; Mengelle, C.; et al. Detection of ganciclovir resistance after valacyclovir-prophylaxis in renal transplant recipients with active cytomegalovirus infection. J. Med. Virol. 2004, 73, 566–573. [Google Scholar] [CrossRef]
- Hantz, S.; Garnier-Geoffroy, F.; Mazeron, M.-C.; Garrigue, I.; Merville, P.; Mengelle, C.; Rostaing, L.; Saint Marcoux, F.; Essig, M.; Rerolle, J.-P.; et al. Drug-resistant cytomegalovirus in transplant recipients: A French cohort study. J. Antimicrob. Chemother. 2010, 65, 2628–2640. [Google Scholar] [CrossRef]
- Lurain, N.S.; Chou, S. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 2010, 23, 689–712. [Google Scholar] [CrossRef]
- Andouard, D.; Mazeron, M.-C.; Ligat, G.; Couvreux, A.; Pouteil-Noble, C.; Cahen, R.; Yasdanpanah, Y.; Deering, M.; Viget, N.; Alain, S.; et al. Contrasting effect of new HCMV pUL54 mutations on antiviral drug susceptibility: Benefits and limits of 3D analysis. Antiviral Res. 2016, 129, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Chemaly, R.F.; Ullmann, A.J.; Stoelben, S.; Richard, M.P.; Bornhäuser, M.; Groth, C.; Einsele, H.; Silverman, M.; Mullane, K.M.; Brown, J.; et al. Letermovir for cytomegalovirus prophylaxis in hematopoietic-cell transplantation. N. Engl. J. Med. 2014, 370, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Lischka, P.; Hewlett, G.; Wunberg, T.; Baumeister, J.; Paulsen, D.; Goldner, T.; Ruebsamen-Schaeff, H.; Zimmermann, H. In vitro and in vivo activities of the novel anticytomegalovirus compound AIC246. Antimicrob. Agents Chemother. 2010, 54, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Goldner, T.; Hewlett, G.; Ettischer, N.; Ruebsamen-Schaeff, H.; Zimmermann, H.; Lischka, P. The novel anticytomegalovirus compound AIC246 (Letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J. Virol. 2011, 85, 10884–10893. [Google Scholar] [CrossRef] [PubMed]
- Melendez, D.P.; Razonable, R.R. Letermovir and inhibitors of the terminase complex: A promising new class of investigational antiviral drugs against human cytomegalovirus. Infect. Drug Resist. 2015, 8, 269–277. [Google Scholar] [PubMed]
- Ligat, G.; Cazal, R.; Hantz, S.; Alain, S. The human cytomegalovirus terminase complex as an antiviral target: A close-up view. FEMS Microbiol. Rev. 2018, 42, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Baines, J.D. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 2006, 80, 5733–5739. [Google Scholar] [CrossRef]
- Baines, J.D.; Cunningham, C.; Nalwanga, D.; Davison, A. The U(L)15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the U(L)15 gene product. J. Virol. 1997, 71, 2666–2673. [Google Scholar] [PubMed]
- Baines, J.D.; Poon, A.P.; Rovnak, J.; Roizman, B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 1994, 68, 8118–8124. [Google Scholar] [PubMed]
- Yang, L.; Yang, Q.; Wang, M.; Jia, R.; Chen, S.; Zhu, D.; Liu, M.; Wu, Y.; Zhao, X.; Zhang, S.; et al. Terminase Large Subunit Provides a New Drug Target for Herpesvirus Treatment. Viruses 2019, 11, 219. [Google Scholar] [CrossRef]
- Champier, G.; Couvreux, A.; Hantz, S.; Rametti, A.; Mazeron, M.-C.; Bouaziz, S.; Denis, F.; Alain, S. Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole D-ribonucleoside activity. Antivir. Ther. 2008, 13, 643–654. [Google Scholar] [PubMed]
- Savva, C.G.W.; Holzenburg, A.; Bogner, E. Insights into the structure of human cytomegalovirus large terminase subunit pUL56. FEBS Lett. 2004, 563, 135–140. [Google Scholar] [CrossRef]
- Hwang, J.-S.; Bogner, E. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J. Biol. Chem. 2002, 277, 6943–6948. [Google Scholar] [CrossRef]
- Ligat, G.; Jacquet, C.; Chou, S.; Couvreux, A.; Alain, S.; Hantz, S. Identification of a short sequence in the HCMV terminase pUL56 essential for interaction with pUL89 subunit. Sci. Rep. 2017, 7, 8796. [Google Scholar] [CrossRef]
- Scholz, B.; Rechter, S.; Drach, J.C.; Townsend, L.B.; Bogner, E. Identification of the ATP-binding site in the terminase subunit pUL56 of human cytomegalovirus. Nucleic Acids Res. 2003, 31, 1426–1433. [Google Scholar] [CrossRef]
- Krosky, P.M.; Underwood, M.R.; Turk, S.R.; Feng, K.W.; Jain, R.K.; Ptak, R.G.; Westerman, A.C.; Biron, K.K.; Townsend, L.B.; Drach, J.C. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 1998, 72, 4721–4728. [Google Scholar]
- Yang, W. Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 2011, 44, 1–93. [Google Scholar] [CrossRef]
- Chevalier, B.S.; Monnat, R.J.; Stoddard, B.L. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat. Struct. Biol. 2001, 8, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pellenz, S.; Ulge, U.; Stoddard, B.L.; Monnat, R.J. Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009, 37, 1650–1662. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, R.; Lambert, A.R.; Mak, A.N.-S.; Jacoby, K.; Dickson, R.J.; Gloor, G.B.; Scharenberg, A.M.; Edgell, D.R.; Stoddard, B.L. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl. Acad. Sci. USA 2011, 108, 13077–13082. [Google Scholar] [CrossRef]
- Silva, G.H.; Dalgaard, J.Z.; Belfort, M.; Van Roey, P. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol. 1999, 286, 1123–1136. [Google Scholar] [CrossRef] [PubMed]
- Chou, S. Rapid In Vitro Evolution of Human Cytomegalovirus UL56 Mutations That Confer Letermovir Resistance. Antimicrob. Agents Chemother. 2015, 59, 6588–6593. [Google Scholar] [CrossRef]
- Chee, M.S.; Bankier, A.T.; Beck, S.; Bohni, R.; Brown, C.M.; Cerny, R.; Horsnell, T.; Hutchison, C.A.; Kouzarides, T.; Martignetti, J.A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 1990, 154, 125–169. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef]
- Borst, E.M.; Hahn, G.; Koszinowski, U.H.; Messerle, M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: A new approach for construction of HCMV mutants. J. Virol. 1999, 73, 8320–8329. [Google Scholar]
- Grishin, A.; Fonfara, I.; Alexeevski, A.; Spirin, S.; Zanegina, O.; Karyagina, A.; Alexeyevsky, D.; Wende, W. Identification of conserved features of LAGLIDADG homing endonucleases. J. Bioinform. Comput. Biol. 2010, 8, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Bogner, E.; Radsak, K.; Stinski, M.F. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 1998, 72, 2259–2264. [Google Scholar] [PubMed]
- Lischka, P.; Michel, D.; Zimmermann, H. Characterization of Cytomegalovirus Breakthrough Events in a Phase 2 Prophylaxis Trial of Letermovir (AIC246, MK 8228). J. Infect. Dis. 2016, 213, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Ligat, G.; Da Re, S.; Alain, S.; Hantz, S. Identification of Amino Acids Essential for Viral Replication in the HCMV Helicase-Primase Complex. Front. Microbiol. 2018, 9, 2483. [Google Scholar] [CrossRef] [Green Version]




| Amino Acids a | Mutation(s) | Mutant HCMV-BAC | Day 4 Post-Tranfection | Day 11 Post-Transfection |
|---|---|---|---|---|
| UL56 WT | / | / | - | ++ |
| LATLNDIERFL | LATLNAIERFL | UL56 D139A | - | + |
| LATLNDIERFL | LATLNDIARFL | UL56 E141A | - | - |
| LATLNDIERFL | AATLNDIERFL | UL56 L134A | - | + |
| LATLNDIERFL | LATANDIERFL | UL56 L137A | - | + |
| LATLNDIERFL | LATLNDAERFL | UL56 I140A | - | + |
| LATLNDIERFL | LATLNDIERFA | UL56 L144A | - | + |
| LATLNDIERFL | AATANDAERFA | UL56 L134A L137A I140A L144A | - | - |
| LATLNDIERFL | LATLNDAERFA | UL56 I140A L144A | - | - |
| LATLNDIERFL | AATLNDIERFA | UL56 L134A L144A | - | - |
| Amino Acids a | Mutation(s) | Mutant HCMV-BAC | Day 4 Post-Transfection | Day 11 Post-Transfection |
|---|---|---|---|---|
| UL56 WT | / | / | - | ++ |
| C-C-NQG-C-H-H | S-C-NQG-C-H-H | UL56 C191S | - | - |
| C-C-NQG-C-H-H | C-S-NQG-C-H-H | UL56 C194S | - | - |
| C-C-NQG-C-H-H | C-C-NQG-S-H-H | UL56 C217S | - | - |
| C-C-NQG-C-H-H | C-C-NQG-C-A-H | UL56 H219A | - | - |
| C-C-NQG-C-H-H | C-C-NQG-C-H-A | UL56 H223A | - | ++ |
| C-C-NQG-C-H-H | C-C-AQG-C-H-H | UL56 N203A | - | - |
| C-C-NQG-C-H-H | C-C-NRG-C-H-H | UL56 Q204R | - | + |
| C-C-NQG-C-H-H | C-C-NQA-C-H-H | UL56 G205A | - | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ligat, G.; Couvreux, A.; Cazal, R.; Alain, S.; Hantz, S. Highlighting of a LAGLIDADG and a Zing Finger Motifs Located in the pUL56 Sequence Crucial for HCMV Replication. Viruses 2019, 11, 1093. https://doi.org/10.3390/v11121093
Ligat G, Couvreux A, Cazal R, Alain S, Hantz S. Highlighting of a LAGLIDADG and a Zing Finger Motifs Located in the pUL56 Sequence Crucial for HCMV Replication. Viruses. 2019; 11(12):1093. https://doi.org/10.3390/v11121093
Chicago/Turabian StyleLigat, Gaëtan, Anthony Couvreux, Raphaël Cazal, Sophie Alain, and Sébastien Hantz. 2019. "Highlighting of a LAGLIDADG and a Zing Finger Motifs Located in the pUL56 Sequence Crucial for HCMV Replication" Viruses 11, no. 12: 1093. https://doi.org/10.3390/v11121093
APA StyleLigat, G., Couvreux, A., Cazal, R., Alain, S., & Hantz, S. (2019). Highlighting of a LAGLIDADG and a Zing Finger Motifs Located in the pUL56 Sequence Crucial for HCMV Replication. Viruses, 11(12), 1093. https://doi.org/10.3390/v11121093