Host Transcription Factors in Hepatitis B Virus RNA Synthesis
Abstract
:1. Introduction
2. The HBV Genome and Control of Viral Replication
3. Transcription Factors
3.1. Ubiquitous Transcription Factors
3.1.1. Nuclear Transcription Factor Y
3.1.2. Nuclear Factor kappa B
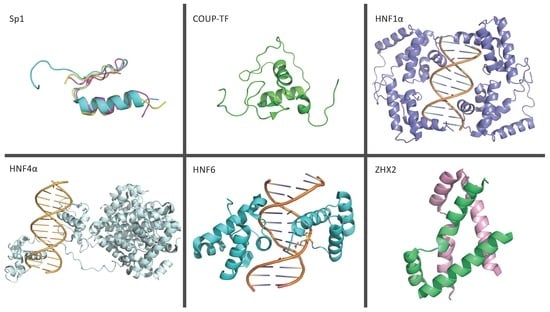
3.1.3. Specificity Protein 1
3.1.4. Chicken Ovalbumin Upstream Promoter Transcription Factor
3.1.5. CCAAT Enhancer Binding Protein Family
3.1.6. Transcription Factor IIB
3.1.7. Zinc-Finger E-Box Binding Homeobox 2
3.1.8. Tumor Protein 53
3.1.9. Regulatory Factor Box 1
3.1.10. Homeobox A10
3.1.11. Octamer Binding Protein 1
3.1.12. Nuclear Respiratory Factor 1
3.1.13. Signal Transducer and Activator of Transcription 1
3.1.14. Signal Transducer and Activator of Transcription 3
3.1.15. Activator Protein 1
3.1.16. Prospero-Related Homeobox Protein 1
3.1.17. TATA Box Protein
3.1.18. Yin Yang 1
3.1.19. Activating Transcription Factor 2
3.1.20. cAMP Response Element-Binding Transcription Factor
3.1.21. Small Heterodimer Partner
3.2. Liver-Enriched Transcription Factors
3.2.1. Hepatocyte Nuclear Factor 1α
3.2.2. Hepatocyte Nuclear Factor 3
3.2.3. Hepatocyte Nuclear Factor 4α
3.2.4. Hepatocyte Nuclear Factor 6
3.2.5. Peroxisome Proliferator-Activated Receptor α
3.2.6. Retinoid X Receptor α
3.2.7. Farsenoid X Receptor α
3.2.8. Zinc Finger and Homeoboxes 2
3.2.9. Krüppel-Like Factor 15
3.2.10. Liver Receptor Homolog 1
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- WHO. Global Hepatitis Report 2017; World Health Organization: Geneva, Switzerland, 2017; p. 7. [Google Scholar]
- Cheng, Z.; Zhi, X.; Sun, G.; Guo, W.; Huang, Y.; Sun, W.; Tian, X.; Zhao, F.; Hu, K. Sodium selenite suppresses hepatitis B virus transcription and replication in human hepatoma cell lines. J. Med. Virol. 2016, 88, 653–663. [Google Scholar] [CrossRef]
- Kim, D.H.; Kang, H.S.; Kim, K.H. Roles of hepatocyte nuclear factors in hepatitis B virus infection. World J. Gastroenterol. 2016, 22, 7017–7029. [Google Scholar] [CrossRef]
- Lu, C.C.; Yen, T.S. Activation of the hepatitis B virus S promoter by transcription factor NF-Y via a CCAAT element. Virology 1996, 225, 387–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moolla, N.; Kew, M.; Arbuthnot, P. Regulatory elements of hepatitis B virus transcription. J. Viral. Hepat. 2002, 9, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.Q.; Duan, Z.; Dai, E.; Zhang, S.; Han, G.; Wang, Y.; Zhang, H.; Zou, H.; Zhu, B.; Zhao, W.; et al. Tenofovir to Prevent Hepatitis B Transmission in Mothers with High Viral Load. N. Engl. J. Med. 2016, 374, 2324–2334. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.J.; Protzer, U. Targeting Innate and Adaptive Immune Responses to Cure Chronic HBV Infection. Gastroenterology 2019, 156, 325–337. [Google Scholar] [CrossRef] [Green Version]
- Tu, T.; Budzinska, M.A.; Vondran, F.W.R.; Shackel, N.A.; Urban, S. Hepatitis B Virus DNA Integration Occurs Early in the Viral Life Cycle in an In Vitro Infection Model via Sodium Taurocholate Cotransporting Polypeptide-Dependent Uptake of Enveloped Virus Particles. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Lucifora, J.; Protzer, U. Attacking hepatitis B virus cccDNA—The holy grail to hepatitis B cure. J. Hepatol. 2016, 64, S41–S48. [Google Scholar] [CrossRef]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppensteiner, H.; Makowska, Z.; Volz, T.; et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014, 343, 1221–1228. [Google Scholar] [CrossRef]
- Beck, J.; Nassal, M. Hepatitis B virus replication. World J. Gastroenterol. 2007, 13, 48–64. [Google Scholar] [CrossRef] [Green Version]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- Quasdorff, M.; Protzer, U. Control of hepatitis B virus at the level of transcription. J. Viral Hepat. 2010, 17, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 2012, 1, e00049. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Saso, W.; Sugiyama, R.; Ishii, K.; Ohki, M.; Nagamori, S.; Suzuki, R.; Aizaki, H.; Ryo, A.; Yun, J.H.; et al. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc. Natl. Acad. Sci. USA 2019, 116, 8487–8492. [Google Scholar] [CrossRef] [Green Version]
- Li, H.C.; Huang, E.Y.; Su, P.Y.; Wu, S.Y.; Yang, C.C.; Lin, Y.S.; Chang, W.C.; Shih, C. Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathog. 2010, 6, e1001162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nassal, M. Hepatitis B viruses: Reverse transcription a different way. Virus Res. 2008, 134, 235–249. [Google Scholar] [CrossRef]
- Seeger, C.; Mason, W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000, 64, 51–68. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.C.; Kao, J.H. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: Molecular mechanisms and clinical significance. Emerg Microbes Infect. 2014, 3, e64. [Google Scholar] [CrossRef]
- Laras, A.; Koskinas, J.; Dimou, E.; Kostamena, A.; Hadziyannis, S.J. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology 2006, 44, 694–702. [Google Scholar] [CrossRef]
- Kao, J.H. Molecular epidemiology of hepatitis B virus. Korean J. Intern. Med. 2011, 26, 255–261. [Google Scholar] [CrossRef]
- Heermann, K.H.; Goldmann, U.; Schwartz, W.; Seyffarth, T.; Baumgarten, H.; Gerlich, W.H. Large surface proteins of hepatitis B virus containing the pre-s sequence. J. Virol. 1984, 52, 396–402. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ou, J.H. Differential regulation of hepatitis B virus gene expression by the Sp1 transcription factor. J. Virol. 2001, 75, 8400–8406. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.J. Hepatitis B: The virus and disease. Hepatology 2009, 49, S13–S21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quarleri, J. Core promoter: A critical region where the hepatitis B virus makes decisions. World J. Gastroenterol. 2014, 20, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Delgermaa, L.; Huang, F.; Oishi, N.; Liu, L.; He, F.; Zhao, L.; Murakami, S. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J. Virol. 2005, 79, 5548–5556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucifora, J.; Arzberger, S.; Durantel, D.; Belloni, L.; Strubin, M.; Levrero, M.; Zoulim, F.; Hantz, O.; Protzer, U. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol. 2011, 55, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S. Hepatitis B virus X protein: A multifunctional viral regulator. J. Gastroenterol. 2001, 36, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.J. Virology: The X-Files of hepatitis B. Nature 2016, 531, 313–314. [Google Scholar] [CrossRef]
- Decorsiere, A.; Mueller, H.; van Breugel, P.C.; Abdul, F.; Gerossier, L.; Beran, R.K.; Livingston, C.M.; Niu, C.; Fletcher, S.P.; Hantz, O.; et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016, 531, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Belloni, L.; Pollicino, T.; De Nicola, F.; Guerrieri, F.; Raffa, G.; Fanciulli, M.; Raimondo, G.; Levrero, M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. USA 2009, 106, 19975–19979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Stephenson, V.; Bremner, W.; Dalton, C.; van Marle, G.; Coffin, C.; Patel, T. Comprehensive Analysis of Hepatitis B Virus Promoter Region Mutations. Viruses 2018, 10, 603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, C.Y.; Choi, B.H.; Park, G.T.; Rho, H.M. Activating transcription factor 2 (ATF2) down-regulates hepatitis B virus X promoter activity by the competition for the activating protein 1 binding site and the formation of the ATF2-Jun heterodimer. J. Biol. Chem. 1997, 272, 16934–16939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauer, U.; Weiss, L.; Lipp, M.; Hofschneider, P.H.; Kekule, A.S. The hepatitis B virus preS2/St transactivator utilizes AP-1 and other transcription factors for transactivation. Hepatology 1994, 19, 23–31. [Google Scholar] [CrossRef]
- Ramji, D.P.; Foka, P. CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem. J. 2002, 365, 561–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.K.; Lim, S.O.; Park, Y.G. Requirement of the cyclic adenosine monophosphate response element-binding protein for hepatitis B virus replication. Hepatology 2008, 48, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Faktor, O.; Budlovsky, S.; Ben-Levy, R.; Shaul, Y. A single element within the hepatitis B virus enhancer binds multiple proteins and responds to multiple stimuli. J. Virol. 1990, 64, 1861–1863. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Hieng, S.; Qian, X.; Costa, R.; Ou, J.H. Regulation of hepatitis B virus ENI enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology 1994, 205, 127–132. [Google Scholar] [CrossRef]
- Yoneyama, M.; Koshiba, S.; Watabe, S.; Harada, T.; Kigawa, T.; Yokoyama, S.; RIKEN Structural Genomics/Proteomics Initiative (RSGI). Solution Structure of the Zinc Finger, C4-type domain of human COUP transcription Factor. Available online: https://www.rcsb.org/structure/2EBL (accessed on 30 March 2019).
- Yang, Q.; Zhang, Q.; Zhang, X.; You, L.; Wang, W.; Liu, W.; Han, Y.; Ma, C.; Xu, W.; Chen, J.; et al. HoxA10 Facilitates SHP-1-Catalyzed Dephosphorylation of p38 MAPK/STAT3 To Repress Hepatitis B Virus Replication by a Feedback Regulatory Mechanism. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Ghosh, S. A glycine-rich region in NF-kappaB p105 functions as a processing signal for the generation of the p50 subunit. Mol. Cell. Biol. 1996, 16, 2248–2254. [Google Scholar] [CrossRef] [Green Version]
- Waris, G.; Huh, K.W.; Siddiqui, A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol. Cell. Biol. 2001, 21, 7721–7730. [Google Scholar] [CrossRef] [Green Version]
- Maity, S.N.; de Crombrugghe, B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998, 23, 174–178. [Google Scholar] [CrossRef]
- Tokusumi, Y.; Zhou, S.; Takada, S. Nuclear respiratory factor 1 plays an essential role in transcriptional initiation from the hepatitis B virus x gene promoter. J. Virol. 2004, 78, 10856–10864. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.B.; Ji, P.; Anish, R.; Jacobson, R.H.; Takada, S. Poly(ADP-ribose) Polymerase 1 Interacts with Nuclear Respiratory Factor 1 (NRF-1) and Plays a Role in NRF-1 Transcriptional Regulation. J. Biol. Chem. 2009, 284, 8621–8632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.X.; Yen, T.S. The ubiquitous transcription factor Oct-1 and the liver-specific factor HNF-1 are both required to activate transcription of a hepatitis B virus promoter. Mol. Cell. Biol. 1991, 11, 1353–1359. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Zhai, J.; Hong, R.; Shan, S.; Kong, Y.; Wen, Y.; Wang, Y.; Liu, J.; Xie, Y. Prospero-related homeobox protein (Prox1) inhibits hepatitis B virus replication through repressing multiple cis regulatory elements. J. Gen. Virol. 2009, 90, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Reith, W.; Ucla, C.; Barras, E.; Gaud, A.; Durand, B.; Herrero-Sanchez, C.; Kobr, M.; Mach, B. RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol. Cell. Biol. 1994, 14, 1230–1244. [Google Scholar] [CrossRef] [Green Version]
- Buckwold, V.E.; Chen, M.; Ou, J.H. Interaction of transcription factors RFX1 and MIBP1 with the gamma motif of the negative regulatory element of the hepatitis B virus core promoter. Virology 1997, 227, 515–518. [Google Scholar] [CrossRef] [Green Version]
- Siegrist, C.A.; Durand, B.; Emery, P.; David, E.; Hearing, P.; Mach, B.; Reith, W. RFX1 is identical to enhancer factor C and functions as a transactivator of the hepatitis B virus enhancer. Mol. Cell. Biol. 1993, 13, 6375–6384. [Google Scholar] [CrossRef] [Green Version]
- Oropeza, C.E.; Li, L.; McLachlan, A. Differential inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by the small heterodimer partner. J. Virol. 2008, 82, 3814–3821. [Google Scholar] [CrossRef] [Green Version]
- Narayan, V.A.; Kriwacki, R.W.; Caradonna, J.P. Structures of zinc finger domains from transcription factor Sp1. Insights into sequence-specific protein-DNA recognition. J. Biol. Chem. 1997, 272, 7801–7809. [Google Scholar] [CrossRef] [Green Version]
- Oka, S.; Shiraishi, Y.; Yoshida, T.; Ohkubo, T.; Sugiura, Y.; Kobayashi, Y. NMR structure of transcription factor Sp1 DNA binding domain. Biochemistry 2004, 43, 16027–16035. [Google Scholar] [CrossRef]
- Wu, M.; Xu, Y.; Lin, S.; Zhang, X.; Xiang, L.; Yuan, Z. Hepatitis B virus polymerase inhibits the interferon-inducible MyD88 promoter by blocking nuclear translocation of Stat1. J. Gen. Virol. 2007, 88, 3260–3269. [Google Scholar] [CrossRef] [PubMed]
- Belloni, L.; Allweiss, L.; Guerrieri, F.; Pediconi, N.; Volz, T.; Pollicino, T.; Petersen, J.; Raimondo, G.; Dandri, M.; Levrero, M. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Investig. 2012, 122, 529–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Zheng, B.; Han, Q.; Zhang, C.; Tian, Z.; Zhang, J. Targeting blockage of STAT3 inhibits hepatitis B virus-related hepatocellular carcinoma. Cancer Biol. Ther. 2016, 17, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Siddiqui, A. Interaction between STAT-3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J. Virol. 2002, 76, 2721–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogomolski-Yahalom, V.; Klein, A.; Greenblat, I.; Haviv, Y.; Tur-Kaspa, R. The TATA-less promoter of hepatitis B virus S gene contains a TBP binding site and an active initiator. Virus Res. 1997, 49, 1–7. [Google Scholar] [CrossRef]
- Ghosh, M.; Elsby, L.M.; Mal, T.K.; Gooding, J.M.; Roberts, S.G.; Ikura, M. Probing Zn2+-binding effects on the zinc-ribbon domain of human general transcription factor TFIIB. Biochem. J. 2004, 378, 317–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Nomura, T.; Cheong, J.; Dorjsuren, D.; Iida, K.; Murakami, S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J. Biol. Chem. 1997, 272, 7132–7139. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kim, H.T.; Yun, Y. Liver-specific enhancer II is the target for the p53-mediated inhibition of hepatitis B viral gene expression. J. Biol. Chem. 1998, 273, 19786–19791. [Google Scholar] [CrossRef] [Green Version]
- Ori, A.; Zauberman, A.; Doitsh, G.; Paran, N.; Oren, M.; Shaul, Y. p53 binds and represses the HBV enhancer: An adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998, 17, 544–553. [Google Scholar] [CrossRef]
- Truant, R.; Antunovic, J.; Greenblatt, J.; Prives, C.; Cromlish, J.A. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J. Virol. 1995, 69, 1851–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakanishi-Matsui, M.; Hayashi, Y.; Kitamura, Y.; Koike, K. Integrated hepatitis B virus DNA preserves the binding sequence of transcription factor Yin and Yang 1 at the virus-cell junction. J. Virol. 2000, 74, 5562–5568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Q.; Li, W.; Ren, J.; Huang, Y.; Huang, Y.; Hu, Q.; Chen, J.; Chen, W. ZEB2 inhibits HBV transcription and replication by targeting its core promoter. Oncotarget 2016, 7, 16003–16011. [Google Scholar] [CrossRef] [PubMed]
- Mouzannar, K.; Fusil, F.; Lacombe, B.; Ollivier, A.; Menard, C.; Lotteau, V.; Cosset, F.L.; Ramiere, C.; Andre, P. Farnesoid X receptor-alpha is a proviral host factor for hepatitis B virus that is inhibited by ligands in vitro and in vivo. FASEB J. 2019, 33, 2472–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramiere, C.; Scholtes, C.; Diaz, O.; Icard, V.; Perrin-Cocon, L.; Trabaud, M.A.; Lotteau, V.; Andre, P. Transactivation of the hepatitis B virus core promoter by the nuclear receptor FXRalpha. J. Virol. 2008, 82, 10832–10840. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Tan, T.; Tian, Y.; Zheng, B.; Ou, J.H.; Huang, E.J.; Yen, T.S. Kruppel-like factor 15 activates hepatitis B virus gene expression and replication. Hepatology 2011, 54, 109–121. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.L.; Liu, Y.Q.; Shan, S.F.; Kong, Y.Y.; Zhou, Q.; Li, M.; Ding, J.P.; Xie, Y.H.; Wang, Y. Molecular mechanism for the potentiation of the transcriptional activity of human liver receptor homolog 1 by steroid receptor coactivator-1. Mol. Endocrinol. 2004, 18, 1887–1905. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.N.; Zhou, Q.; Kong, Y.Y.; Li, M.; Viollet, B.; Xie, Y.H.; Wang, Y. LRH-1/hB1F and HNF1 synergistically up-regulate hepatitis B virus gene transcription and DNA replication. Cell Res. 2003, 13, 451–458. [Google Scholar] [CrossRef]
- Li, J.; Xu, Z.; Zheng, Y.; Johnson, D.L.; Ou, J.H. Regulation of hepatocyte nuclear factor 1 activity by wild-type and mutant hepatitis B virus X proteins. J. Virol. 2002, 76, 5875–5881. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Li, J.; Ou, J.H. Regulation of hepatitis B virus core promoter by transcription factors HNF1 and HNF4 and the viral X protein. J. Virol. 2004, 78, 6908–6914. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xie, Y.; Wu, X.; Kong, Y.; Wang, Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology 1995, 214, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Raney, A.K.; Zhang, P.; McLachlan, A. Regulation of transcription from the hepatitis B virus large surface antigen promoter by hepatocyte nuclear factor 3. J. Virol. 1995, 69, 3265–3272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Chen, M.; Yen, T.S.; Ou, J.H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol. Cell. Biol. 1993, 13, 443–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.Y.; Kim, H.J.; Park, C.; So, H.S.; Park, R.K.; Kim, H.C. Impact of Nucleotide Mutations at the HNF3- and HNF4-Binding Sites in Enhancer 1 on Viral Replication in Patients with Chronic Hepatitis B Virus Infection. Gut Liver 2013, 7, 569–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, Y.; Chen, E.; Liu, C.; Huang, F.; Zhou, T.; He, F.; Liu, L.; Liu, F.; Tang, H. The correlation of hepatocyte nuclear factor 4 alpha and 3 beta with hepatitis B virus replication in the liver of chronic hepatitis B patients. J. Viral Hepat. 2009, 16, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.; He, J.; Liu, X.; Gao, G.; Liu, D.; Cui, L.; Yu, G.; Yu, W.; Chen, Y.; Guo, D. Inhibition of hepatitis B virus gene expression and replication by hepatocyte nuclear factor 6. J. Virol. 2015, 89, 4345–4355. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Ma, Y.; Liu, M.; Yan, L.; Tang, H. Peroxisome Proliferators Activated Receptor (PPAR) agonists activate hepatitis B virus replication in vivo. Virol. J. 2017, 14, 96. [Google Scholar] [CrossRef] [Green Version]
- Huan, B.; Kosovsky, M.J.; Siddiqui, A. Retinoid X receptor alpha transactivates the hepatitis B virus enhancer 1 element by forming a heterodimeric complex with the peroxisome proliferator-activated receptor. J. Virol. 1995, 69, 547–551. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Raney, A.K.; McLachlan, A. Replication of the wild type and a natural hepatitis B virus nucleocapsid promoter variant is differentially regulated by nuclear hormone receptors in cell culture. J. Virol. 2001, 75, 8937–8948. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wu, Z.; Tan, S.; Wang, Z.; Lin, Q.; Li, X.; Song, X.; Liu, Y.; Song, Y.; Zhang, J.; et al. Tumor suppressor ZHX2 restricts hepatitis B virus replication via epigenetic and non-epigenetic manners. Antivir. Res. 2018, 153, 114–123. [Google Scholar] [CrossRef]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Agostino, S.; Strano, S.; Emiliozzi, V.; Zerbini, V.; Mottolese, M.; Sacchi, A.; Blandino, G.; Piaggio, G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer cell 2006, 10, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolfini, D.; Gatta, R.; Mantovani, R. NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Foo, N.C.; Yen, T.S. Activation of promoters for cellular lipogenic genes by hepatitis B virus large surface protein. Virology 2000, 269, 420–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, K.; Karin, M. NF-kappaB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol 2018, 18, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.W.; Rey, F.A.; Sodeoka, M.; Verdine, G.L.; Harrison, S.C. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature 1995, 373, 311–317. [Google Scholar] [CrossRef]
- Berkowitz, B.; Huang, D.B.; Chen-Park, F.E.; Sigler, P.B.; Ghosh, G. The x-ray crystal structure of the NF-kappa B p50.p65 heterodimer bound to the interferon beta -kappa B site. J. Biol. Chem. 2002, 277, 24694–24700. [Google Scholar] [CrossRef] [Green Version]
- Tak, P.P.; Firestein, G.S. NF-kappaB: A key role in inflammatory diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Warner, N.; Ryan, K.; Selleck, L.; Colledge, D.; Rodgers, S.; Li, K.; Revill, P.; Locarnini, S. The hepatitis B e antigen suppresses IL-1beta-mediated NF-kappaB activation in hepatocytes. J. Viral Hepat. 2011, 18, e499–e507. [Google Scholar] [CrossRef]
- Hiscott, J.; Kwon, H.; Genin, P. Hostile takeovers: Viral appropriation of the NF-kappaB pathway. J. Clin. Investig. 2001, 107, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Cao, H.; Lu, J.; Shu, X.; Xiong, X.; Hong, X.; Xu, Q.; Zhu, H.; Li, G.; Shen, G. Interleukin-32 expression induced by hepatitis B virus protein X is mediated through activation of NF-kappaB. Mol. Immunol. 2011, 48, 1573–1577. [Google Scholar] [CrossRef] [PubMed]
- Kaczynski, J.; Cook, T.; Urrutia, R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003, 4, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, B.Y.; Tran, K.; Ku, T.K.; Crowe, D.L. Regulation of ERK1 gene expression by coactivator proteins. Biochem. J. 2005, 392, 589–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, A.R.; Black, J.D.; Azizkhan-Clifford, J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 2001, 188, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.Y.; Khachigian, L.M. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell. Biol. 2009, 29, 2483–2488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizcaino, C.; Mansilla, S.; Portugal, J. Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol. Ther. 2015, 152, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Alfano, C.; Magrinelli, E.; Harb, K.; Studer, M. The nuclear receptors COUP-TF: A long-lasting experience in forebrain assembly. Cell Mol. Life Sci. 2014, 71, 43–62. [Google Scholar] [CrossRef]
- Tsai, S.Y.; Tsai, M.J. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): Coming of age. Endocr Rev. 1997, 18, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Vivanco Ruiz, M.M.; Bugge, T.H.; Hirschmann, P.; Stunnenberg, H.G. Functional characterization of a natural retinoic acid responsive element. EMBO J. 1991, 10, 3829–3838. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Umesono, K.; Heyman, R.A.; Mangelsdorf, D.J.; Dyck, J.A.; Evans, R.M. Retinoid X receptor-COUP-TF interactions modulate retinoic acid signaling. Proc. Natl. Acad. Sci. USA 1992, 89, 1448–1452. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Tsai, S.Y.; Tsai, M.J. COUP-TF an orphan member of the steroid/thyroid hormone receptor superfamily. Trends Endocrinol Metab 1994, 5, 234–239. [Google Scholar] [CrossRef]
- Pereira, F.A.; Tsai, M.J.; Tsai, S.Y. COUP-TF orphan nuclear receptors in development and differentiation. Cell Mol. Life Sci. 2000, 57, 1388–1398. [Google Scholar] [CrossRef]
- Tripodi, M.; Filosa, A.; Armentano, M.; Studer, M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development 2004, 131, 6119–6129. [Google Scholar] [CrossRef] [Green Version]
- Garcia, A.D.; Ostapchuk, P.; Hearing, P. Functional interaction of nuclear factors EF-C, HNF-4, and RXR alpha with hepatitis B virus enhancer I. J. Virol. 1993, 67, 3940–3950. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Ito, M.; Sun, S.; Chida, T.; Nakashima, K.; Suzuki, T. LUC7L3/CROP inhibits replication of hepatitis B virus via suppressing enhancer II/basal core promoter activity. Sci. Rep. 2016, 6, 36741. [Google Scholar] [CrossRef]
- Yu, X.; Mertz, J.E. Distinct modes of regulation of transcription of hepatitis B virus by the nuclear receptors HNF4alpha and COUP-TF1. J. Virol. 2003, 77, 2489–2499. [Google Scholar] [CrossRef] [Green Version]
- Lekstrom-Himes, J.; Xanthopoulos, K.G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998, 273, 28545–28548. [Google Scholar] [CrossRef] [Green Version]
- Takiguchi, M. The C/EBP family of transcription factors in the liver and other organs. Int. J. Exp. Pathol. 1998, 79, 369–391. [Google Scholar] [CrossRef]
- Wang, W.; Xia, X.; Mao, L.; Wang, S. The CCAAT/Enhancer-Binding Protein Family: Its Roles in MDSC Expansion and Function. Front. Immunol. 2019, 10, 1804. [Google Scholar] [CrossRef]
- Choi, B.H.; Park, G.T.; Rho, H.M. Interaction of hepatitis B viral X protein and CCAAT/ enhancer-binding protein alpha synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J. Biol. Chem. 1999, 274, 2858–2865. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Cabrera, M.; Letovsky, J.; Hu, K.Q.; Siddiqui, A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: Trans-activation and repression by CCAAT/enhancer binding protein. Proc. Natl. Acad. Sci. USA 1990, 87, 5069–5073. [Google Scholar] [CrossRef] [Green Version]
- Raney, A.K.; McLachlan, A. Characterization of the hepatitis B virus large surface antigen promoter Sp1 binding site. Virology 1995, 208, 399–404. [Google Scholar] [CrossRef] [Green Version]
- Bock, C.T.; Kubicka, S.; Manns, M.P.; Trautwein, C. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology 1999, 29, 1236–1247. [Google Scholar] [CrossRef]
- Sarkar, N.; Chakravarty, R. Hepatitis B Virus Infection, MicroRNAs and Liver Disease. Int. J. Mol. Sci. 2015, 16, 17746–17762. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, N.; Panigrahi, R.; Pal, A.; Biswas, A.; Singh, S.P.; Kar, S.K.; Bandopadhyay, M.; Das, D.; Saha, D.; Kanda, T.; et al. Expression of microRNA-155 correlates positively with the expression of Toll-like receptor 7 and modulates hepatitis B virus via C/EBP-beta in hepatocytes. J. Viral Hepat. 2015, 22, 817–827. [Google Scholar] [CrossRef]
- Sainsbury, S.; Niesser, J.; Cramer, P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 2013, 493, 437–440. [Google Scholar] [CrossRef] [Green Version]
- Haviv, I.; Shamay, M.; Doitsh, G.; Shaul, Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol. Cell. Biol. 1998, 18, 1562–1569. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Huang, F.; Chen, L.; Chen, E.; Bai, L.; Cheng, X.; He, M.; Tang, H. RPB5-Mediating Protein Suppresses Hepatitis B Virus (HBV) Transcription and Replication by Counteracting the Transcriptional Activation of Hepatitis B virus X Protein in HBV Replication Mouse Model. Jundishapur J. Microbiol. 2015, 8, e21936. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhang, H.; Ye, L. Effects of hepatitis B virus X protein on the development of liver cancer. J. Lab. Clin. Med. 2006, 147, 58–66. [Google Scholar] [CrossRef]
- Verschueren, K.; Remacle, J.E.; Collart, C.; Kraft, H.; Baker, B.S.; Tylzanowski, P.; Nelles, L.; Wuytens, G.; Su, M.T.; Bodmer, R.; et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5’-CACCT sequences in candidate target genes. J. Biol. Chem. 1999, 274, 20489–20498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postigo, A.A.; Depp, J.L.; Taylor, J.J.; Kroll, K.L. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003, 22, 2453–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Putte, T.; Maruhashi, M.; Francis, A.; Nelles, L.; Kondoh, H.; Huylebroeck, D.; Higashi, Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am. J. Hum. Genet. 2003, 72, 465–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Putte, T.; Francis, A.; Nelles, L.; van Grunsven, L.A.; Huylebroeck, D. Neural crest-specific removal of Zfhx1b in mouse leads to a wide range of neurocristopathies reminiscent of Mowat-Wilson syndrome. Hum. Mol. Genet. 2007, 16, 1423–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epifanova, E.; Babaev, A.; Newman, A.G.; Tarabykin, V. Role of Zeb2/Sip1 in neuronal development. Brain Res. 2019, 1705, 24–31. [Google Scholar] [CrossRef]
- Remacle, J.E.; Kraft, H.; Lerchner, W.; Wuytens, G.; Collart, C.; Verschueren, K.; Smith, J.C.; Huylebroeck, D. New mode of DNA binding of multi-zinc finger transcription factors: DeltaEF1 family members bind with two hands to two target sites. EMBO J. 1999, 18, 5073–5084. [Google Scholar] [CrossRef] [Green Version]
- Long, J.; Zuo, D.; Park, M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J. Biol. Chem. 2005, 280, 35477–35489. [Google Scholar] [CrossRef] [Green Version]
- Kamada, R.; Toguchi, Y.; Nomura, T.; Imagawa, T.; Sakaguchi, K. Tetramer formation of tumor suppressor protein p53: Structure, function, and applications. Biopolymers 2016, 106, 598–612. [Google Scholar] [CrossRef]
- Vieler, M.; Sanyal, S. p53 Isoforms and Their Implications in Cancer. Cancers 2018, 10, 288. [Google Scholar] [CrossRef] [Green Version]
- Kearns, S.; Lurz, R.; Orlova, E.V.; Okorokov, A.L. Two p53 tetramers bind one consensus DNA response element. Nucleic Acids Res. 2016, 44, 6185–6199. [Google Scholar] [CrossRef] [Green Version]
- Kitayner, M.; Rozenberg, H.; Kessler, N.; Rabinovich, D.; Shaulov, L.; Haran, T.E.; Shakked, Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell 2006, 22, 741–753. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.H.; Won, J.; Yun, Y. Through induction of juxtaposition and tyrosine kinase activity of Jak1, X-gene product of hepatitis B virus stimulates Ras and the transcriptional activation through AP-1, NF-kappaB, and SRE enhancers. Biochem. Biophys. Res. Commun. 2001, 286, 886–894. [Google Scholar] [CrossRef]
- Chan, C.; Thurnherr, T.; Wang, J.; Gallart-Palau, X.; Sze, S.K.; Rozen, S.; Lee, C.G. Global re-wiring of p53 transcription regulation by the hepatitis B virus X protein. Mol. Oncol. 2016, 10, 1183–1195. [Google Scholar] [CrossRef]
- Katan-Khaykovich, Y.; Shaul, Y. Nuclear import and DNA-binding activity of RFX1. Evidence for an autoinhibitory mechanism. Eur. J. Biochem. 2001, 268, 3108–3116. [Google Scholar] [CrossRef] [Green Version]
- Gajiwala, K.S.; Chen, H.; Cornille, F.; Roques, B.P.; Reith, W.; Mach, B.; Burley, S.K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 2000, 403, 916–921. [Google Scholar] [CrossRef]
- Julien, L.; Chassagne, J.; Peccate, C.; Lorain, S.; Pietri-Rouxel, F.; Danos, O.; Benkhelifa-Ziyyat, S. RFX1 and RFX3 Transcription Factors Interact with the D Sequence of Adeno-Associated Virus Inverted Terminal Repeat and Regulate AAV Transduction. Sci. Rep. 2018, 8, 210. [Google Scholar] [CrossRef]
- Zanatta, A.; Rocha, A.M.; Carvalho, F.M.; Pereira, R.M.; Taylor, H.S.; Motta, E.L.; Baracat, E.C.; Serafini, P.C. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: A review. J. Assist. Reprod Genet. 2010, 27, 701–710. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Taylor, H.S. The Role of Hox Genes in Female Reproductive Tract Development, Adult Function, and Fertility. Cold Spring Harbor. Perspect. Med. 2015, 6, a023002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, F.; Yang, X.; He, Y.; Wang, H.; Aili, A.; Ding, Y. Aberrant endometrial DNA methylome of homeobox A10 and catechol-O-methyltransferase in endometriosis. J. Assist. Reprod. Genet. 2017, 34, 409–415. [Google Scholar] [CrossRef] [Green Version]
- Benson, G.V.; Nguyen, T.H.; Maas, R.L. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol. Cell. Biol. 1995, 15, 1591–1601. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Peter, W.; Ciesiolka, T.; Gruss, P.; Scholer, H.R. Mouse Oct-1 contains a composite homeodomain of human Oct-1 and Oct-2. Nucleic Acids Res. 1993, 21, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemler, I.; Schaffner, W. Octamer transcription factors and the cell type-specificity of immunoglobulin gene expression. FASEB J. 1990, 4, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Herr, W.; Cleary, M.A. The POU domain: Versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995, 9, 1679–1693. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.S.; Kim, L.K.; Lee, G.R.; Flavell, R.A. Role of OCT-1 and partner proteins in T cell differentiation. Biochim. Biophys. Acta 2016, 1859, 825–831. [Google Scholar] [CrossRef]
- Tantin, D.; Schild-Poulter, C.; Wang, V.; Hache, R.J.; Sharp, P.A. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res. 2005, 65, 10750–10758. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Arreguin, K.; Tantin, D. The Oct1 transcription factor and epithelial malignancies: Old protein learns new tricks. Biochim. Biophys. Acta 2016, 1859, 792–804. [Google Scholar] [CrossRef] [Green Version]
- Antunovic, J.; Lemieux, N.; Cromlish, J.A. The 17 kDa HBx protein encoded by hepatitis B virus interacts with the activation domains of Oct-1, and functions as a coactivator in the activation and repression of a human U6 promoter. Cell Mol. Biol Res. 1993, 39, 463–482. [Google Scholar]
- Scarpulla, R.C. Nuclear control of respiratory gene expression in mammalian cells. J. Cell Biochem. 2006, 97, 673–683. [Google Scholar] [CrossRef]
- Niu, N.; Li, Z.; Zhu, M.; Sun, H.; Yang, J.; Xu, S.; Zhao, W.; Song, R. Effects of nuclear respiratory factor1 on apoptosis and mitochondrial dysfunction induced by cobalt chloride in H9C2 cells. Mol. Med. Rep. 2019, 19, 2153–2163. [Google Scholar] [CrossRef] [Green Version]
- Morrish, F.; Giedt, C.; Hockenbery, D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003, 17, 240–255. [Google Scholar] [CrossRef] [Green Version]
- Virbasius, C.A.; Virbasius, J.V.; Scarpulla, R.C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993, 7, 2431–2445. [Google Scholar] [CrossRef]
- Gugneja, S.; Scarpulla, R.C. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J. Biol. Chem. 1997, 272, 18732–18739. [Google Scholar] [CrossRef] [Green Version]
- Meissl, K.; Macho-Maschler, S.; Muller, M.; Strobl, B. The good and the bad faces of STAT1 in solid tumours. Cytokine 2017, 89, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Mogensen, T.H. IRF and STAT Transcription Factors - From Basic Biology to Roles in Infection, Protective Immunity, and Primary Immunodeficiencies. Front. Immunol. 2018, 9, 3047. [Google Scholar] [CrossRef]
- Roy, B.; Zuo, Z.; Stormo, G.D. Quantitative specificity of STAT1 and several variants. Nucleic Acids Res. 2017, 45, 8199–8207. [Google Scholar] [CrossRef] [Green Version]
- Ehret, G.B.; Reichenbach, P.; Schindler, U.; Horvath, C.M.; Fritz, S.; Nabholz, M.; Bucher, P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 2001, 276, 6675–6688. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Tang, H. Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein. Protein Cell 2010, 1, 1106–1117. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Yu, J.; Guo, H.; Song, H.; Chen, S. Upregulation of survivin by leptin/STAT3 signaling in MCF-7 cells. Biochem. Biophys. Res. Commun. 2008, 368, 1–5. [Google Scholar] [CrossRef]
- Tsai, Y.T.; Su, Y.H.; Fang, S.S.; Huang, T.N.; Qiu, Y.; Jou, Y.S.; Shih, H.M.; Kung, H.J.; Chen, R.H. Etk, a Btk family tyrosine kinase, mediates cellular transformation by linking Src to STAT3 activation. Mol. Cell. Biol. 2000, 20, 2043–2054. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Huang, C.; Wang, X.J.; Xin, D.E.; Wang, L.S.; Zou, Q.C.; Zhang, Y.S.; Tan, M.D.; Wang, Y.M.; Zhao, T.C.; et al. Lysyl Oxidase 3 Is a Dual-Specificity Enzyme Involved in STAT3 Deacetylation and Deacetylimination Modulation. Mol. Cell 2017, 65, 296–309. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Cherukuri, P.; Luo, J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J. Biol. Chem. 2005, 280, 11528–11534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garces de Los Fayos Alonso, I.; Liang, H.C.; Turner, S.D.; Lagger, S.; Merkel, O.; Kenner, L. The Role of Activator Protein-1 (AP-1) Family Members in CD30-Positive Lymphomas. Cancers 2018, 10, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papoudou-Bai, A.; Hatzimichael, E.; Barbouti, A.; Kanavaros, P. Expression patterns of the activator protein-1 (AP-1) family members in lymphoid neoplasms. Clin. Exp. Med. 2017, 17, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Trop-Steinberg, S.; Azar, Y. AP-1 Expression and its Clinical Relevance in Immune Disorders and Cancer. Am. J. Med. Sci. 2017, 353, 474–483. [Google Scholar] [CrossRef]
- Angel, P.; Imagawa, M.; Chiu, R.; Stein, B.; Imbra, R.J.; Rahmsdorf, H.J.; Jonat, C.; Herrlich, P.; Karin, M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 1987, 49, 729–739. [Google Scholar] [CrossRef]
- Kang, L.J.; Choi, Y.J.; Lee, S.G. Stimulation of TRAF6/TAK1 degradation and inhibition of JNK/AP-1 signalling by ginsenoside Rg3 attenuates hepatitis B virus replication. Int. J. Biochem. Cell Biol. 2013, 45, 2612–2621. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kanai, F.; Ichimura, T.; Tateishi, K.; Asaoka, Y.; Guleng, B.; Jazag, A.; Ohta, M.; Imamura, J.; Ikenoue, T.; et al. The hepatitis B virus X protein enhances AP-1 activation through interaction with Jab1. Oncogene 2006, 25, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Ryu, C.J.; Hong, H.J. Hepatitis B virus preS1 functions as a transcriptional activation domain. J. Gen. Virol 1997, 78 Pt 5, 1083–1086. [Google Scholar] [CrossRef] [Green Version]
- Elsir, T.; Smits, A.; Lindstrom, M.S.; Nister, M. Transcription factor PROX1: Its role in development and cancer. Cancer Metastasis Rev. 2012, 31, 793–805. [Google Scholar] [CrossRef]
- Takeda, Y.; Jetten, A.M. Prospero-related homeobox 1 (Prox1) functions as a novel modulator of retinoic acid-related orphan receptors alpha- and gamma-mediated transactivation. Nucleic Acids Res. 2013, 41, 6992–7008. [Google Scholar] [CrossRef] [Green Version]
- Stergiopoulos, A.; Elkouris, M.; Politis, P.K. Prospero-related homeobox 1 (Prox1) at the crossroads of diverse pathways during adult neural fate specification. Front. Cell Neurosci. 2014, 8, 454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audette, D.S.; Anand, D.; So, T.; Rubenstein, T.B.; Lachke, S.A.; Lovicu, F.J.; Duncan, M.K. Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development 2016, 143, 318–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Gao, D.M.; Jiang, Q.F.; Zhou, Q.; Kong, Y.Y.; Wang, Y.; Xie, Y.H. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-alpha-hydroxylase gene. Mol. Endocrinol. 2004, 18, 2424–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, A.; Sinn, E.; Yamamoto, T.; Wang, J.; Roy, A.; Horikoshi, M.; Roeder, R.G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature 1990, 346, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, D.B.; Chen, H.; Halay, E.D.; Hoffman, A.; Roeder, R.G.; Burley, S.K. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc. Natl. Acad. Sci. USA 1996, 93, 4862–4867. [Google Scholar] [CrossRef] [Green Version]
- Nikolov, D.B.; Hu, S.H.; Lin, J.; Gasch, A.; Hoffmann, A.; Horikoshi, M.; Chua, N.H.; Roeder, R.G.; Burley, S.K. Crystal structure of TFIID TATA-box binding protein. Nature 1992, 360, 40–46. [Google Scholar] [CrossRef]
- Qadri, I.; Maguire, H.F.; Siddiqui, A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc. Natl. Acad. Sci. USA 1995, 92, 1003–1007. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.C.; Chen, J.J.; Yang, P.C. Multifunctional transcription factor YY1: A therapeutic target in human cancer? Expert Opin. Ther. Targets 2006, 10, 253–266. [Google Scholar] [CrossRef]
- Galloway, N.R.; Ball, K.F.; Stiff, T.; Wall, N.R. Yin Yang 1 (YY1): Regulation of Survivin and Its Role In Invasion and Metastasis. Crit. Rev. Oncog. 2017, 22, 23–36. [Google Scholar] [CrossRef]
- Shi, Y.; Lee, J.S.; Galvin, K.M. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1997, 1332, F49–F66. [Google Scholar] [CrossRef]
- Zhang, X.; Hou, J.; Lu, M. Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front. Genet. 2013, 4, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, Y.; Kitamura, Y.; Nakanishi, M.; Koike, K. The binding site of transcription factor YY1 is required for intramolecular recombination between terminally repeated sequences of linear replicative hepatitis B virus DNA. J. Virol. 2000, 74, 9471–9478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, J.; Zheng, Y.; Guo, X.; Mo, J.; Xie, X.; Xiong, Y.; Liu, Y.; Wu, K.; Wu, J. Hepatitis B virus replication and sex-determining region Y box 4 production are tightly controlled by a novel positive feedback mechanism. Sci. Rep. 2015, 5, 10066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Li, Y.J.; Bian, A.H.; Zuo, H.B.; Zhu, T.W.; Ji, S.X.; Kong, F.; Yin, D.Q.; Wang, C.B.; Wang, Z.F.; et al. The regulatory role of activating transcription factor 2 in inflammation. Mediators Inflamm. 2014, 2014, 950472. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, H.; Schiltz, L.; Chiu, R.; Itakura, K.; Taira, K.; Nakatani, Y.; Yokoyama, K.K. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 2000, 405, 195–200. [Google Scholar] [CrossRef]
- Bhoumik, A.; Takahashi, S.; Breitweiser, W.; Shiloh, Y.; Jones, N.; Ronai, Z. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol. Cell 2005, 18, 577–587. [Google Scholar] [CrossRef] [Green Version]
- Nagadoi, A.; Nakazawa, K.; Uda, H.; Okuno, K.; Maekawa, T.; Ishii, S.; Nishimura, Y. Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain. J. Mol. Biol. 1999, 287, 593–607. [Google Scholar] [CrossRef]
- Newell, C.L.; Deisseroth, A.B.; Lopez-Berestein, G. Interaction of nuclear proteins with an AP-1/CRE-like promoter sequence in the human TNF-alpha gene. J. Leukoc Biol 1994, 56, 27–35. [Google Scholar] [CrossRef]
- Alberini, C.M. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Frank, D.A. CREB in the pathophysiology of cancer: Implications for targeting transcription factors for cancer therapy. Clin. Cancer Res. 2009, 15, 2583–2587. [Google Scholar] [CrossRef] [Green Version]
- Herzig, S.; Long, F.; Jhala, U.S.; Hedrick, S.; Quinn, R.; Bauer, A.; Rudolph, D.; Schutz, G.; Yoon, C.; Puigserver, P.; et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 2001, 413, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Adya, N.; Zhao, L.J.; Huang, W.; Boros, I.; Giam, C.Z. Expansion of CREB’s DNA recognition specificity by Tax results from interaction with Ala-Ala-Arg at positions 282-284 near the conserved DNA-binding domain of CREB. Proc. Natl. Acad. Sci. USA 1994, 91, 5642–5646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H.M.; Gao, W.W.; Chan, C.P.; Cheng, Y.; Chaudhary, V.; Deng, J.J.; Yuen, K.S.; Wong, C.M.; Ng, I.O.; Kok, K.H.; et al. Requirement of CRTC1 coactivator for hepatitis B virus transcription. Nucleic Acids Res. 2014, 42, 12455–12468. [Google Scholar] [CrossRef] [Green Version]
- Yuk, J.M.; Jin, H.S.; Jo, E.K. Small Heterodimer Partner and Innate Immune Regulation. Endocrinol. Metab. 2016, 31, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Wang, L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res. 2008, 36, 5727–5735. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hagedorn, C.H.; Wang, L. Role of nuclear receptor SHP in metabolism and cancer. Biochim. Biophys. Acta 2011, 1812, 893–908. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Xiao, Z.; Kanamaluru, D.; Min, G.; Yau, P.M.; Veenstra, T.D.; Ellis, E.; Strom, S.; Suino-Powell, K.; Xu, H.E.; et al. Bile acid signaling pathways increase stability of Small Heterodimer Partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 2009, 23, 986–996. [Google Scholar] [CrossRef] [Green Version]
- Reese, V.C.; Moore, D.D.; McLachlan, A. Limited effects of bile acids and small heterodimer partner on hepatitis B virus biosynthesis in vivo. J. Virol. 2012, 86, 2760–2768. [Google Scholar] [CrossRef] [Green Version]
- Chi, Y.I.; Frantz, J.D.; Oh, B.C.; Hansen, L.; Dhe-Paganon, S.; Shoelson, S.E. Diabetes mutations delineate an atypical POU domain in HNF-1alpha. Mol. Cell 2002, 10, 1129–1137. [Google Scholar] [CrossRef]
- Chandra, V.; Huang, P.; Potluri, N.; Wu, D.; Kim, Y.; Rastinejad, F. Multidomain integration in the structure of the HNF-4alpha nuclear receptor complex. Nature 2013, 495, 394–398. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, M.; Bonvin, A.M. 3D-DART: A DNA structure modelling server. Nucleic Acids Res. 2009, 37, W235–W239. [Google Scholar] [CrossRef] [PubMed]
- Iyaguchi, D.; Yao, M.; Watanabe, N.; Nishihira, J.; Tanaka, I. DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6. Structure 2007, 15, 75–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, L.E.; Ren, J.; Nettleship, J.E.; Folkers, G.E.; Owens, R.J.; Stammers, D.K. Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains. BMC Struct. Biol. 2010, 10, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zong, L.; Sureau, C.; Barker, L.; Wands, J.R.; Tong, S. Unusual Features of Sodium Taurocholate Cotransporting Polypeptide as a Hepatitis B Virus Receptor. J. Virol. 2016, 90, 8302–8313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Falth, M.; Stindt, J.; Koniger, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146, 1070–1083. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Qi, Y.; Peng, B.; Li, W. NTCP-Reconstituted In Vitro HBV Infection System. Methods Mol. Biol. 2017, 1540, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, S.; Liang, W. Bona fide receptor for hepatitis B and D viral infections: Mechanism, research models and molecular drug targets. Emerg. Microbes Infect. 2018, 7, 134. [Google Scholar] [CrossRef]
- Singh, V.; Singla, S.K.; Jha, V.; Puri, V.; Puri, S. Hepatocyte nuclear factor-1beta: A regulator of kidney development and cystogenesis. Indian J. Nephrol. 2015, 25, 70–76. [Google Scholar] [CrossRef]
- Lau, H.H.; Ng, N.H.J.; Loo, L.S.W.; Jasmen, J.B.; Teo, A.K.K. The molecular functions of hepatocyte nuclear factors - In and beyond the liver. J. Hepatol. 2018, 68, 1033–1048. [Google Scholar] [CrossRef] [Green Version]
- De Simone, V.; De Magistris, L.; Lazzaro, D.; Gerstner, J.; Monaci, P.; Nicosia, A.; Cortese, R. LFB3, a heterodimer-forming homeoprotein of the LFB1 family, is expressed in specialized epithelia. EMBO J. 1991, 10, 1435–1443. [Google Scholar] [CrossRef]
- Rey-Campos, J.; Chouard, T.; Yaniv, M.; Cereghini, S. vHNF1 is a homeoprotein that activates transcription and forms heterodimers with HNF1. EMBO J. 1991, 10, 1445–1457. [Google Scholar] [CrossRef]
- Odom, D.T.; Zizlsperger, N.; Gordon, D.B.; Bell, G.W.; Rinaldi, N.J.; Murray, H.L.; Volkert, T.L.; Schreiber, J.; Rolfe, P.A.; Gifford, D.K.; et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 2004, 303, 1378–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servitja, J.M.; Pignatelli, M.; Maestro, M.A.; Cardalda, C.; Boj, S.F.; Lozano, J.; Blanco, E.; Lafuente, A.; McCarthy, M.I.; Sumoy, L.; et al. Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol. Cell. Biol. 2009, 29, 2945–2959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Wnuck Lipinski, K.; Sattler, K.; Peters, S.; Weske, S.; Keul, P.; Klump, H.; Heusch, G.; Gothert, J.R.; Levkau, B. Hepatocyte Nuclear Factor 1A Is a Cell-Intrinsic Transcription Factor Required for B Cell Differentiation and Development in Mice. J. Immunol. 2016, 196, 1655–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.X.; Li, M.; Wu, X.; Wang, Y.; Li, Z.P. HNF1 is critical for the liver-specific function of HBV enhancer II. Res. Virol. 1998, 149, 99–108. [Google Scholar] [CrossRef]
- Bar-Yishay, I.; Shaul, Y.; Shlomai, A. Hepatocyte metabolic signalling pathways and regulation of hepatitis B virus expression. Liver Int. 2011, 31, 282–290. [Google Scholar] [CrossRef]
- Lin, J.; Gu, C.; Shen, Z.; Liu, Y.; Wang, W.; Tao, S.; Cui, X.; Liu, J.; Xie, Y. Hepatocyte nuclear factor 1alpha downregulates HBV gene expression and replication by activating the NF-kappaB signaling pathway. PLoS ONE 2017, 12, e0174017. [Google Scholar] [CrossRef]
- Li, J.; Dantas Machado, A.C.; Guo, M.; Sagendorf, J.M.; Zhou, Z.; Jiang, L.; Chen, X.; Wu, D.; Qu, L.; Chen, Z.; et al. Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site. Biochemistry 2017, 56, 3745–3753. [Google Scholar] [CrossRef]
- Sund, N.J.; Ang, S.L.; Sackett, S.D.; Shen, W.; Daigle, N.; Magnuson, M.A.; Kaestner, K.H. Hepatocyte nuclear factor 3beta (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol. Cell. Biol. 2000, 20, 5175–5183. [Google Scholar] [CrossRef] [Green Version]
- Sund, N.J.; Vatamaniuk, M.Z.; Casey, M.; Ang, S.L.; Magnuson, M.A.; Stoffers, D.A.; Matschinsky, F.M.; Kaestner, K.H. Tissue-specific deletion of Foxa2 in pancreatic beta cells results in hyperinsulinemic hypoglycemia. Genes Dev. 2001, 15, 1706–1715. [Google Scholar] [CrossRef] [Green Version]
- Wolfrum, C.; Stoffel, M. Coactivation of Foxa2 through Pgc-1beta promotes liver fatty acid oxidation and triglyceride/VLDL secretion. Cell Metab. 2006, 3, 99–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ori, A.; Shaul, Y. Hepatitis B virus enhancer binds and is activated by the Hepatocyte nuclear factor 3. Virology 1995, 207, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; McLachlan, A. Mechanisms of inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by hepatocyte nuclear factor 3beta. J. Virol. 2002, 76, 8572–8581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirillo, L.A.; Lin, F.R.; Cuesta, I.; Friedman, D.; Jarnik, M.; Zaret, K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 2002, 9, 279–289. [Google Scholar] [CrossRef]
- Gallastegui, N.; Mackinnon, J.A.; Fletterick, R.J.; Estebanez-Perpina, E. Advances in our structural understanding of orphan nuclear receptors. Trends Biochem. Sci. 2015, 40, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Drewes, T.; Senkel, S.; Holewa, B.; Ryffel, G.U. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol. Cell. Biol. 1996, 16, 925–931. [Google Scholar] [CrossRef] [Green Version]
- Babeu, J.P.; Boudreau, F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J. Gastroenterol. 2014, 20, 22–30. [Google Scholar] [CrossRef]
- Hwang-Verslues, W.W.; Sladek, F.M. HNF4alpha—Role in drug metabolism and potential drug target? Curr. Opin. Pharmacol. 2010, 10, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Vasoya, R.P.; Toke, N.H.; Parthasarathy, A.; Luo, S.; Chiles, E.; Flores, J.; Gao, N.; Bonder, E.M.; Su, X.; et al. HNF4 Regulates Fatty Acid Oxidation and is Required for Renewal of Intestinal Stem Cells in Mice. Gastroenterology 2019. [Google Scholar] [CrossRef]
- Garrison, W.D.; Battle, M.A.; Yang, C.; Kaestner, K.H.; Sladek, F.M.; Duncan, S.A. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology 2006, 130, 1207–1220. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Cheng, X.; Li, Y.; Valdez, K.; Chen, W.; Liang, T.J. Hepatitis B Virus Deregulates the Cell Cycle To Promote Viral Replication and a Premalignant Phenotype. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, F.; Chen, E.Q.; Liu, L.; Zhou, T.Y.; Liu, C.; Cheng, X.; Liu, F.J.; Tang, H. Inhibition of hepatitis B Virus replication by hepatocyte nuclear factor 4-alpha specific short hairpin RNA. Liver Int. 2012, 32, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Yeh, S.H.; Lin, W.H.; Yeh, K.H.; Yuan, Q.; Xia, N.S.; Chen, D.S.; Chen, P.J. Estrogen receptor alpha represses transcription of HBV genes via interaction with hepatocyte nuclear factor 4alpha. Gastroenterology 2012, 142, 989–998.e4. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Yan, H.; Rausa, F.M., 3rd; Costa, R.H.; Liao, X. Structure of the hepatocyte nuclear factor 6alpha and its interaction with DNA. J. Biol. Chem. 2004, 279, 33928–33936. [Google Scholar] [CrossRef] [Green Version]
- Iyaguchi, D.; Yao, M.; Watanabe, N.; Nishihira, J.; Tanaka, I. Crystallization and preliminary X-ray studies of the DNA-binding domain of hepatocyte nuclear factor-6alpha complexed with DNA. Protein Pept Lett. 2006, 13, 531–533. [Google Scholar] [CrossRef]
- Zhang, H.; Ables, E.T.; Pope, C.F.; Washington, M.K.; Hipkens, S.; Means, A.L.; Path, G.; Seufert, J.; Costa, R.H.; Leiter, A.B.; et al. Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech. Dev. 2009, 126, 958–973. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Fang, B.; Damle, M.; Guan, D.; Li, Z.; Kim, Y.H.; Gannon, M.; Lazar, M.A. HNF6 and Rev-erbalpha integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev. 2016, 30, 1636–1644. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.W.; Wang, D.M.; Hu, Y.; Tang, Y.N.; Shi, W.W.; Guo, X.J.; Song, J.G. Hepatocyte nuclear factor 6 suppresses the migration and invasive growth of lung cancer cells through p53 and the inhibition of epithelial-mesenchymal transition. J. Biol. Chem. 2013, 288, 31206–31216. [Google Scholar] [CrossRef] [Green Version]
- Jacquemin, P.; Durviaux, S.M.; Jensen, J.; Godfraind, C.; Gradwohl, G.; Guillemot, F.; Madsen, O.D.; Carmeliet, P.; Dewerchin, M.; Collen, D.; et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol. Cell. Biol. 2000, 20, 4445–4454. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Wang, M.; Gannon, M.; Holterman, A. Growth Hormone Mediates Its Protective Effect in Hepatic Apoptosis through Hnf6. PLoS ONE 2016, 11, e0167085. [Google Scholar] [CrossRef] [Green Version]
- Lahuna, O.; Fernandez, L.; Karlsson, H.; Maiter, D.; Lemaigre, F.P.; Rousseau, G.G.; Gustafsson, J.; Mode, A. Expression of hepatocyte nuclear factor 6 in rat liver is sex-dependent and regulated by growth hormone. Proc. Natl. Acad. Sci. USA 1997, 94, 12309–12313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 2011, 1812, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patsouris, D.; Mandard, S.; Voshol, P.J.; Escher, P.; Tan, N.S.; Havekes, L.M.; Koenig, W.; Marz, W.; Tafuri, S.; Wahli, W.; et al. PPARalpha governs glycerol metabolism. J. Clin. Investig. 2004, 114, 94–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burri, L.; Thoresen, G.H.; Berge, R.K. The Role of PPARalpha Activation in Liver and Muscle. PPAR Res. 2010, 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra, V.; Huang, P.; Hamuro, Y.; Raghuram, S.; Wang, Y.; Burris, T.P.; Rastinejad, F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature 2008, 456, 350–356. [Google Scholar] [CrossRef]
- Dubuquoy, L.; Louvet, A.; Hollebecque, A.; Mathurin, P.; Dharancy, S. Peroxisome proliferator-activated receptors in HBV-related infection. PPAR Res. 2009, 2009, 145124. [Google Scholar] [CrossRef] [Green Version]
- Raney, A.K.; Kline, E.F.; Tang, H.; McLachlan, A. Transcription and replication of a natural hepatitis B virus nucleocapsid promoter variant is regulated in vivo by peroxisome proliferators. Virology 2001, 289, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Li, Y.; Huang, C.; Ying, L.; Xue, J.; Wu, H.; Chen, Z.; Yang, Z. Resveratrol enhances HBV replication through activating Sirt1-PGC-1alpha-PPARalpha pathway. Sci. Rep. 2016, 6, 24744. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Wang, X.; Ding, X.; Li, Y.; Zhang, X.; Xie, P.; Yang, J.; Wang, S. MicroRNA-141 represses HBV replication by targeting PPARA. PLoS ONE 2012, 7, e34165. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.K.; Hoffmann, B.; Tran, P.B.; Graupner, G.; Pfahl, M. Retinoid X receptor is an auxiliary protein for thyroid hormone and retinoic acid receptors. Nature 1992, 355, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Kliewer, S.A.; Provencal, J.; Wright, P.E.; Evans, R.M. Structure of the retinoid X receptor alpha DNA binding domain: A helix required for homodimeric DNA binding. Science 1993, 260, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Sun, Y.; Tian, J.; He, W.; Xu, G.; Jing, Z.; Li, W. Silencing Retinoid X Receptor Alpha Expression Enhances Early-Stage Hepatitis B Virus Infection In Cell Cultures. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.D.; Chen, W.D.; Moore, D.D.; Huang, W. FXR: A metabolic regulator and cell protector. Cell Res. 2008, 18, 1087–1095. [Google Scholar] [CrossRef] [Green Version]
- Pineda Torra, I.; Claudel, T.; Duval, C.; Kosykh, V.; Fruchart, J.C.; Staels, B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol. Endocrinol. 2003, 17, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Lu, Y.; Li, X.Y. Farnesoid X receptor: A master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacol. Sin. 2015, 36, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Claudel, T.; Staels, B.; Kuipers, F. The Farnesoid X receptor: A molecular link between bile acid and lipid and glucose metabolism. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 2020–2030. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.D.; Chen, W.D.; Wang, X.; Lou, G.; Liu, N.; Lin, M.; Forman, B.M.; Huang, W. Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice. Hepatology 2012, 56, 2336–2343. [Google Scholar] [CrossRef] [Green Version]
- Mangelsdorf, D.J.; Evans, R.M. The RXR heterodimers and orphan receptors. Cell 1995, 83, 841–850. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.M.; Hart, S.N.; Kong, B.; Fang, J.; Zhong, X.B.; Guo, G.L. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 2010, 51, 1410–1419. [Google Scholar] [CrossRef] [Green Version]
- Curtil, C.; Enache, L.S.; Radreau, P.; Dron, A.G.; Scholtes, C.; Deloire, A.; Roche, D.; Lotteau, V.; Andre, P.; Ramiere, C. The metabolic sensors FXRalpha, PGC-1alpha, and SIRT1 cooperatively regulate hepatitis B virus transcription. FASEB J. 2014, 28, 1454–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhang, E.; Ma, Z.; Pei, R.; Jiang, M.; Schlaak, J.F.; Roggendorf, M.; Lu, M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology 2011, 53, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Kawata, H.; Yamada, K.; Shou, Z.; Mizutani, T.; Yazawa, T.; Yoshino, M.; Sekiguchi, T.; Kajitani, T.; Miyamoto, K. Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem. J. 2003, 373, 747–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Ma, D.; Ji, C. Zinc fingers and homeoboxes family in human diseases. Cancer Gene Ther. 2015, 22, 223–226. [Google Scholar] [CrossRef]
- Shen, H.; Luan, F.; Liu, H.; Gao, L.; Liang, X.; Zhang, L.; Sun, W.; Ma, C. ZHX2 is a repressor of alpha-fetoprotein expression in human hepatoma cell lines. J. Cell Mol. Med. 2008, 12, 2772–2780. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Creasy, K.T.; Purnell, J.; Peterson, M.L.; Spear, B.T. Zhx2 (zinc fingers and homeoboxes 2) regulates major urinary protein gene expression in the mouse liver. J. Biol. Chem. 2017, 292, 6765–6774. [Google Scholar] [CrossRef] [Green Version]
- Wan, F.; Gao, L.; Lu, Y.; Ma, H.; Wang, H.; Liang, X.; Wang, Y.; Ma, C. Proliferation and osteo/odontogenic differentiation of stem cells from apical papilla regulated by Zinc fingers and homeoboxes 2: An in vitro study. Biochem. Biophys. Res. Commun. 2016, 469, 599–605. [Google Scholar] [CrossRef]
- Kwon, R.J.; Kim, Y.H.; Jeong, D.C.; Han, M.E.; Kim, J.Y.; Liu, L.; Jung, J.S.; Oh, S.O. Expression and prognostic significance of zinc fingers and homeoboxes family members in renal cell carcinoma. PLoS ONE 2017, 12, e0171036. [Google Scholar] [CrossRef]
- Kawata, H.; Yamada, K.; Shou, Z.; Mizutani, T.; Miyamoto, K. The mouse zinc-fingers and homeoboxes (ZHX) family; ZHX2 forms a heterodimer with ZHX3. Gene 2003, 323, 133–140. [Google Scholar] [CrossRef]
- Zhou, S.J.; Deng, Y.L.; Liang, H.F.; Jaoude, J.C.; Liu, F.Y. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017, 24, 1577–1587. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Tan, S.; Wu, Z.; Xu, L.; Wang, Z.; Xu, Y.; Wang, T.; Gao, C.; Gong, Y.; Liang, X.; et al. HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int. J. Cancer 2018, 143, 3120–3130. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.; Wang, B.; Orihuela, Y.; Hong, E.G.; Fisch, S.; Haldar, S.; Cline, G.W.; Kim, J.K.; Peroni, O.D.; Kahn, B.B.; et al. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 2007, 5, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takashima, M.; Ogawa, W.; Hayashi, K.; Inoue, H.; Kinoshita, S.; Okamoto, Y.; Sakaue, H.; Wataoka, Y.; Emi, A.; Senga, Y.; et al. Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes 2010, 59, 1608–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matoba, K.; Lu, Y.; Zhang, R.; Chen, E.R.; Sangwung, P.; Wang, B.; Prosdocimo, D.A.; Jain, M.K. Adipose KLF15 Controls Lipid Handling to Adapt to Nutrient Availability. Cell Rep. 2017, 21, 3129–3140. [Google Scholar] [CrossRef]
- Pei, J.; Grishin, N.V. C2H2 zinc finger proteins of the SP/KLF, Wilms tumor, EGR, Huckebein, and Klumpfuss families in metazoans and beyond. Gene 2015, 573, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Marcos, P.J.; Auwerx, J.; Schoonjans, K. Emerging actions of the nuclear receptor LRH-1 in the gut. Biochim. Biophys. Acta 2011, 1812, 947–955. [Google Scholar] [CrossRef] [Green Version]
- Fayard, E.; Auwerx, J.; Schoonjans, K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004, 14, 250–260. [Google Scholar] [CrossRef]
- Yumoto, F.; Nguyen, P.; Sablin, E.P.; Baxter, J.D.; Webb, P.; Fletterick, R.J. Structural basis of coactivation of liver receptor homolog-1 by beta-catenin. Proc. Natl. Acad. Sci. USA 2012, 109, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Holmstrom, S.R.; Deering, T.; Swift, G.H.; Poelwijk, F.J.; Mangelsdorf, D.J.; Kliewer, S.A.; MacDonald, R.J. LRH-1 and PTF1-L coregulate an exocrine pancreas-specific transcriptional network for digestive function. Genes Dev. 2011, 25, 1674–1679. [Google Scholar] [CrossRef] [Green Version]
- Stein, S.; Oosterveer, M.H.; Mataki, C.; Xu, P.; Lemos, V.; Havinga, R.; Dittner, C.; Ryu, D.; Menzies, K.J.; Wang, X.; et al. SUMOylation-dependent LRH-1/PROX1 interaction promotes atherosclerosis by decreasing hepatic reverse cholesterol transport. Cell Metab. 2014, 20, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Pollicino, T.; Belloni, L.; Raffa, G.; Pediconi, N.; Squadrito, G.; Raimondo, G.; Levrero, M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 2006, 130, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Urban, S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Moyo, B.; Bloom, K.; Scott, T.; Ely, A.; Arbuthnot, P. Advances with using CRISPR/Cas-mediated gene editing to treat infections with hepatitis B virus and hepatitis C virus. Virus Res. 2018, 244, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Schiwon, M.; Ehrke-Schulz, E.; Oswald, A.; Bergmann, T.; Michler, T.; Protzer, U.; Ehrhardt, A. One-Vector System for Multiplexed CRISPR/Cas9 against Hepatitis B Virus cccDNA Utilizing High-Capacity Adenoviral Vectors. Mol. Ther Nucleic Acids 2018, 12, 242–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Cheng, G.; Mahato, R.I. RNAi for treating hepatitis B viral infection. Pharm. Res. 2008, 25, 72–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flisiak, R.; Jaroszewicz, J.; Lucejko, M. siRNA drug development against hepatitis B virus infection. Expert Opin. Biol. Ther. 2018, 18, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y. Preclinical and Clinical Advances of GalNAc-Decorated Nucleic Acid Therapeutics. Mol. Ther Nucleic Acids 2017, 6, 116–132. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Kim, E.S.; Guo, H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology 2017, 66, 2066–2077. [Google Scholar] [CrossRef]
- Nehme, Z.; Pasquereau, S.; Herbein, G. Control of viral infections by epigenetic-targeted therapy. Clin. Epigenetics 2019, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Tropberger, P.; Mercier, A.; Robinson, M.; Zhong, W.; Ganem, D.E.; Holdorf, M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc. Natl. Acad. Sci. USA 2015, 112, E5715–E5724. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.T.; Guo, H. Metabolism and function of hepatitis B virus cccDNA: Implications for the development of cccDNA-targeting antiviral therapeutics. Antivir. Res. 2015, 122, 91–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonvin, A.M.; Boelens, R.; Kaptein, R. NMR analysis of protein interactions. Curr. Opin. Chem. Biol. 2005, 9, 501–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y. A glimpse of structural biology through X-ray crystallography. Cell 2014, 159, 995–1014. [Google Scholar] [CrossRef] [Green Version]
- Venien-Bryan, C.; Li, Z.; Vuillard, L.; Boutin, J.A. Cryo-electron microscopy and X-ray crystallography: Complementary approaches to structural biology and drug discovery. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 174–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, T.R.; Chojnowski, G.; Koul, A.; McKenna, S.A.; Bujnicki, J.M. Structural studies of RNA-protein complexes: A hybrid approach involving hydrodynamics, scattering, and computational methods. Methods 2017, 118–119, 146–162. [Google Scholar] [CrossRef] [PubMed]



| Protein | Effect on Transcription | Site of HBV Genome Interaction | References |
|---|---|---|---|
| Ubiquitous Transcription Factors | |||
| Activating transcription Factor 2 (ATF2) | Inhibits transcription | Binds ENI region | [33] |
| Activator Protein 1 (AP-1) | Enhances transcription | Binds PreS2/S region | [34] |
| CCAAT Enhancer Binding Protein (C/EBP) | Enhances transcription | Binds PreS1 and ENII promoter regions | [5,13,35] |
| cAMP response element-binding transcription factor (CREB) | Enhances transcription | Binds ENI region | [36,37] |
| Chicken Ovalbumin Upstream Promoter Transcription Factor (COUP-TF) | Inhibits transcription | Binds ENI, core and PreS2 regions | [13,38,39] |
| Homeobox A10 (HOXA10) | Inhibits transcription | Binds ENI and X promoter region | [40] |
| Nuclear Factor kappa B (NF-κB) | Inhibits transcription | Initiate an immune response or is activated by HBx mediated oxidative stress | [41,42] |
| Nuclear Transcription Factor Y (NF-Y) | Enhances transcription | Binds PreS2/S regions | [4,41,43] |
| Nuclear Respiratory Factor 1 (NRF1) | Enhanced transcription | Binds X promoter region | [44,45] |
| Octamer binding protein 1 (Oct1) | Enhances transcription | Binds PreS1 region | [46] |
| Prospero-related homeobox protein 1 (PROX1) | Inhibits transcription | Binds ENI, ENII and PreS1 regions | [47] |
| Regulatory Factor Box 1 (RFX1) | Inhibits transcription | Binds ENI and core promoter regions | [48,49,50] |
| Small heterodimer partner (SHP) d | Inhibits transcription | Binds ENI, ENII, core and X promoter regions | [51] |
| Specificity protein 1 (Sp1) | Enhances transcription | Binds PreC, ENII and PreS2 promoter regions | [13,23,52,53] |
| Signal Transducer and Activator of Transcription 1 (STAT1) | Inhibits transcription | Activates an immune response a binds ENI/X promoter region | [54,55] |
| Signal Transducer and Activator of Transcription 3 (STAT3) | Enhances transcription | Binds ENI region | [56,57] |
| TATA Box Protein (TBP) | Enhances transcription | Binds PreS2/S region | [58] |
| Transcription Factor IIB (TFIIB) | Enhances transcription | Binds X promoter region | [59,60] |
| Tumor Protein 53 (p53) | Inhibits transcription | Binds ENII and X promoter regions | [61,62,63] |
| Yin Yang 1 (YY1) | Inhibits transcription b | Binds Upstream of Direct repeat region 1 | [64] |
| Zinc-finger E-box Binding Homeobox 2 (ZEB2) | Inhibits transcription | Binds core promoter region | [65] |
| Hepatocyte-specific Transcription Factors | |||
| Farsenoid X Receptor α (FXRα) | Enhances transcription | Binds ENII and core promoter | [66,67] |
| Krüppel-like Factor 15 (KLF15) | Enhances transcription | Binds PreS2/S and core promoter | [68] |
| Liver receptor homolog 1 (LRH-1) | Enhances transcription | Binds ENII region | [69,70] |
| Hepatocyte Nuclear Factor 1α (HNF1α) | Enhances and inhibits transcription c | Binds PreS1, ENII, core and X promoter regions | [46,71,72] |
| Hepatocyte Nuclear Factor 3 (HNF3) | Enhances transcription | Binds ENII and PreS1 promoter regions | [38,73,74] |
| Hepatocyte Nuclear Factor 4 α (HNF4α) | Enhances transcription | Binds PreS1, core and ENII promoter regions | [75,76,77] |
| Hepatocyte Nuclear Factor 6 (HNF6) | Inhibits transcription | Binds PreS1/S2 promoter regions | [78] |
| Peroxisome Proliferator-Activated Receptor α (PPARα) | Enhances transcription | Binds ENI, Core and PreS2/S promoter regions | [79,80] |
| Retinoid X Receptor α (RXRα) | Enhances transcription | Binds ENI, Core and S promoter | [80,81] |
| Zinc Finger and Homeoboxes 2 (ZHX2) | Inhibits transcription | Binds X, core and PreS2 promoter | [82] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turton, K.L.; Meier-Stephenson, V.; Badmalia, M.D.; Coffin, C.S.; Patel, T.R. Host Transcription Factors in Hepatitis B Virus RNA Synthesis. Viruses 2020, 12, 160. https://doi.org/10.3390/v12020160
Turton KL, Meier-Stephenson V, Badmalia MD, Coffin CS, Patel TR. Host Transcription Factors in Hepatitis B Virus RNA Synthesis. Viruses. 2020; 12(2):160. https://doi.org/10.3390/v12020160
Chicago/Turabian StyleTurton, Kristi L., Vanessa Meier-Stephenson, Maulik D. Badmalia, Carla S. Coffin, and Trushar R. Patel. 2020. "Host Transcription Factors in Hepatitis B Virus RNA Synthesis" Viruses 12, no. 2: 160. https://doi.org/10.3390/v12020160
APA StyleTurton, K. L., Meier-Stephenson, V., Badmalia, M. D., Coffin, C. S., & Patel, T. R. (2020). Host Transcription Factors in Hepatitis B Virus RNA Synthesis. Viruses, 12(2), 160. https://doi.org/10.3390/v12020160