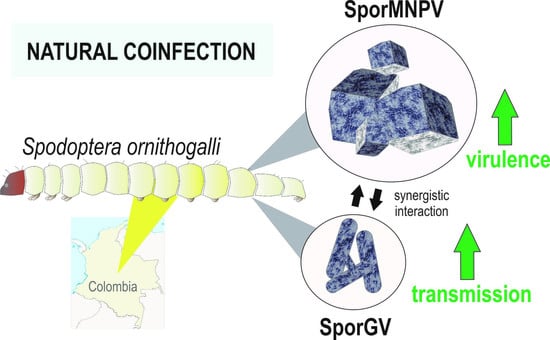
Natural Coinfection between Novel Species of Baculoviruses in Spodoptera ornithogalli Larvae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Virus Production, Purification, and Quantification
2.3. Electron Microscopy
2.4. Genome Sequencing and Analysis
2.5. Biological Activity of Natural Mixed Viruses
2.6. Biological Activity of Purified Viruses and Artificial Mixtures
3. Results
3.1. SporNPV and SporGV
3.1.1. Ultrastructural Studies and Infection Signs
3.1.2. Molecular Characterization
3.2. SporNPV and SporGV Biological Activity
Insecticidal Activity of Wild Viral Mixture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brito, R.; Specht, A.; Gonçalves, G.L.; Moreira, G.R.P.; Carneiro, E.; Santos, F.L.; Roque-Specht, V.F.; Mielke, O.H.H.; Casagrande, M.M. Spodoptera marima: A new synonym of Spodoptera ornithogalli (Lepidoptera: Noctuidae), with notes on adult morphology, host plant use and genetic variation along its geographic range. Neotrop. Entomol. 2019, 48, 433–448. [Google Scholar] [CrossRef]
- Vega-Chávez, J.L.; Hernández, J.M.; Gonzalez-Hernández, H.; Morales-Maldonado, E.R.; Cepeda, Y.A.A. First Report of Spodoptera ornithogalli1 in Agave salmiana. Southwest. Entomol. 2021, 46, 271–274. [Google Scholar] [CrossRef]
- Sermeño-Chicas, J.M.; Rodríguez-Urrutia, E.A.; Joyce, A.L.; Parada-Berrios, F.A.; Pérez, D.; Quintanilla-Quintanilla, J.R. Gusano cuerudo Spodoptera ornithogalli (Guenée 1852) (Lepidoptera: Noctuidae) en cacao Theobroma cacao L. en El Salvador. Rev. Minerva 2017, 1, 62–72. [Google Scholar]
- Sosa-Gómez, D.R. Microbial control of soybean pest insects and mites. In Microbial Control of Insect and Mite Pests; Elsevier: Amsterdam, The Netherlands, 2017; pp. 199–208. [Google Scholar]
- Thézé, J.; Lopez-Vaamonde, C.; Cory, J.S.; Herniou, E.A. Biodiversity, evolution and ecological specialization of baculoviruses: A treasure trove for future applied research. Viruses 2018, 10, 366. [Google Scholar] [CrossRef] [Green Version]
- Jehle, J.A.; Lange, M.; Wang, H.; Hu, Z.; Wang, Y.; Hauschild, R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 2006, 346, 180–193. [Google Scholar] [CrossRef] [Green Version]
- Hostetter, D.L.; Puttler, B.; Mcintosh, A.H.; Pinnell, R.E. A Nuclear Polyhedrosis Virus of the Yellowstriped Armyworm (Lepidoptera: Noctuidae). Environ. Entomol. 1990, 19, 1190–1195. [Google Scholar] [CrossRef]
- Young, S.Y. Effect of nuclear polyhedrosis virus infection in Spodoptera ornithogalli larvae on post larval stages and dissemination by adults. J. Invertebr. Pathol. 1990, 55, 69–75. [Google Scholar] [CrossRef]
- Barrera, G.; Cuartas, P.; Gómez, J.; Guevara, J.; Villamizar, L. Spodoptera ornithogalli nucleopolyhedrovirus: Preliminary study of Colombian isolate. Spodoptera Ornithogalli Nucl. Prelim. Study Colomb. Isol. 2011, 66, 453–456. [Google Scholar]
- Wennmann, J.T.; Alletti, G.G.; Jehle, J.A. The genome sequence of Agrotis segetum nucleopolyhedrovirus B (AgseNPV-B) reveals a new baculovirus species within the Agrotis baculovirus complex. Virus Genes 2015, 50, 260–276. [Google Scholar] [CrossRef]
- Koppenhöfer, A.M.; Kaya, H.K. Additive and Synergistic Interaction between Entomopathogenic Nematodes andBacillus thuringiensisfor Scarab Grub Control. Biol. Control 1997, 8, 131–137. [Google Scholar] [CrossRef]
- Otálora, P.C.; Rivero, L.V. Interacciones de los Virus Entomopatógenos y su Efecto sobre la Actividad Biológica. Rev. Fac. Ciencias Básicas 2011, 7, 220–239. [Google Scholar]
- Cuartas-Otálora, P.E.; Gómez-Valderrama, J.A.; Ramos, A.E.; Barrera-Cubillos, G.P.; Villamizar-Rivero, L.F. Bio-Insecticidal Potential of Nucleopolyhedrovirus and Granulovirus Mixtures to Control the Fall Armyworm Spodoptera frugiperda (JE Smith, 1797) (Lepidoptera: Noctuidae). Viruses 2019, 11, 684. [Google Scholar] [CrossRef] [Green Version]
- Hughes, P.R.; Wood, H.A. A synchronous peroral technique for the bioassay of insect viruses. J. Invertebr. Pathol. 1981, 37, 154–159. [Google Scholar] [CrossRef]
- Caballero, P.; Zuidema, D.; Santiago-Alvarez, C.; Vlak, J.M. Biochemical and biological characterization of four isolates of Spodoptera exigua nuclear polyhedrosis virus. Biocontrol Sci. Technol. 1992, 2, 145–157. [Google Scholar] [CrossRef]
- Sporleder, M.; Kroschel, J.; Huber, J.; Lagnaoui, A. An improved method to determine the biological activity (LC50) of the granulovirus PoGV in its host Phthorimaea operculella. Entomol. Exp. Appl. 2005, 116, 191–197. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Böhme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.-A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef] [Green Version]
- Fickett, J.W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982, 10, 5303–5318. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Minh, B.Q.; Hahn, M.W.; Lanfear, R. New methods to calculate concordance factors for phylogenomic datasets. Mol. Biol. Evol. 2020, 37, 2727–2733. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Keilwagen, J.; Jehle, J.A. Baculovirus Kimura two-parameter species demarcation criterion is confirmed by the distances of 38 core gene nucleotide sequences. J. Gen. Virol. 2018, 99, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [Green Version]
- Crawley, M.J. GLIM for Ecologists; Blackwell Scientific Publications: Oxford, UK, 1993. [Google Scholar]
- LeOra, S. Polo Plus, Probit and Logit Analysis: User’s Guide Computer Program, Version 2.0; LeOra Software: Berkeley, CA, USA, 2007. [Google Scholar]
- Jehle, J.A.; Blissard, G.W.; Bonning, B.C.; Cory, J.S.; Herniou, E.A.; Rohrmann, G.F.; Theilmann, D.A.; Thiem, S.M.; Vlak, J.M. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006, 151, 1257–1266. [Google Scholar] [CrossRef]
- Bideshi, D.K.; Bigot, Y.; Federici, B.A. Molecular characterization and phylogenetic analysis of the Harrisina brillians granulovirus granulin gene. Arch. Virol. 2000, 145, 1933–1945. [Google Scholar] [CrossRef]
- Garavaglia, M.J.; Miele, S.A.B.; Iserte, J.A.; Belaich, M.N.; Ghiringhelli, P.D. The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J. Virol. 2012, 86, 12069–12079. [Google Scholar] [CrossRef] [Green Version]
- Javed, M.A.; Biswas, S.; Willis, L.G.; Harris, S.; Pritchard, C.; van Oers, M.M.; Donly, B.C.; Erlandson, M.A.; Hegedus, D.D.; Theilmann, D.A. Autographa californica multiple nucleopolyhedrovirus AC83 is a per os infectivity factor (PIF) protein required for occlusion-derived virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the PIF complex in ODV envelopes. J. Virol. 2017, 91, e02115-16. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Choi, J.Y.; Roh, J.Y.; Liu, Q.; Tao, X.Y.; Park, J.B.; Kim, J.S.; Je, Y.H. Genomic sequence analysis of granulovirus isolated from the tobacco cutworm, Spodoptera litura. PLoS ONE 2011, 6, e28163. [Google Scholar] [CrossRef] [Green Version]
- Ishimwe, E.; Hodgson, J.J.; Passarelli, A.L. Expression of the Cydia pomonella granulovirus matrix metalloprotease enhances Autographa californica multiple nucleopolyhedrovirus virulence and can partially substitute for viral cathepsin. Virology 2015, 481, 166–178. [Google Scholar] [CrossRef] [Green Version]
- Popham, H.J.R.; Rowley, D.L.; Harrison, R.L. Differential insecticidal properties of Spodoptera frugiperda multiple nucleopolyhedrovirus isolates against corn-strain and rice-strain fall armyworm, and genomic analysis of three isolates. J. Invertebr. Pathol. 2021, 183, 107561. [Google Scholar] [CrossRef]
- Cory, J.S.; Myers, J.H. The Ecology and Evolution of Insect Baculoviruses. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 239–272. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.F.; Fang, J.C.; Wang, J.P.; Zhong, W.F.; Liu, B.S. Interaction of Xestia c-nigrum granulovirus with peritrophic matrix and Spodoptera litura nucleopolyhedrovirus in Spodoptera litura. J. Econ. Entomol. 2007, 100, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, T.; Shimojo, E.; Mori, M.; Kaido, M.; Furusawa, I.; Miyata, S.; Sano, Y.; Matsumoto, T.; Hashimoto, Y.; Granados, R.R. Enhancement of baculovirus infection in Spodoptera exigua (Lepidoptera: Noctuidae) larvae with Autographa californica nucleopolyhedrovirus or Nicotiana tabacum engineered with a granulovirus enhancin gene. Appl. Entomol. Zool. 2000, 35, 163–170. [Google Scholar] [CrossRef] [Green Version]
- DaPalma, T.; Doonan, B.P.; Trager, N.M.; Kasman, L.M. A systematic approach to virus–virus interactions. Virus Res. 2010, 149, 1–9. [Google Scholar] [CrossRef]
- Hoover, K.; Humphries, M.A.; Gendron, A.R.; Slavicek, J.M. Impact of viral enhancin genes on potency of Lymantria dispar multiple nucleopolyhedrovirus in L. dispar following disruption of the peritrophic matrix. J. Invertebr. Pathol. 2010, 104, 150–152. [Google Scholar] [CrossRef]
- Slavicek, J.M. Baculovirus enhancins and their role in viral pathogenicity. In Molecular Virology; IntechOpen: London, UK, 2012; pp. 147–168. [Google Scholar] [CrossRef]
- Rao, R.; Fiandra, L.; Giordana, B.; De Eguileor, M.; Congiu, T.; Burlini, N.; Arciello, S.; Corrado, G.; Pennacchio, F. AcMNPV ChiA protein disrupts the peritrophic membrane and alters midgut physiology of Bombyx mori larvae. Insect Biochem. Mol. Biol. 2004, 34, 1205–1213. [Google Scholar] [CrossRef]
- Cory, J.S. Insect virus transmission: Different routes to persistence. Curr. Opin. Insect Sci. 2015, 8, 130–135. [Google Scholar] [CrossRef]
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Becnel, J.J.; Krell, P.J.; van Oers, M.M.; Mowery, J.D.; Bauchan, G.R. ICTV virus taxonomy profile: Baculoviridae. J. Gen. Virol. 2018, 99, 1185–1186. [Google Scholar] [CrossRef]
- Alletti, G.G.; Eigenbrod, M.; Carstens, E.B.; Kleespies, R.G.; Jehle, J.A. The genome sequence of Agrotis segetum granulovirus, isolate AgseGV-DA, reveals a new Betabaculovirus species of a slow killing granulovirus. J. Invertebr. Pathol. 2017, 146, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Ogembo, J.G.; Chaeychomsri, S.; Caoili, B.L.; Ikeda, M.; Kobayashi, M. Susceptibility of newly established cell lines from Helicoverpa armigera to homologous and heterologous nucleopolyhedroviruses. J. Insect Biotechnol. Sericology 2008, 77, 1_25–1_34. [Google Scholar]
- Harrison, R.L.; Popham, H.J.R. Genomic sequence analysis of a granulovirus isolated from the Old World bollworm, Helicoverpa armigera. Virus Genes 2008, 36, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Rowley, D.L.; Mowery, J.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The complete genome sequence of a second distinct betabaculovirus from the true armyworm, Mythimna unipuncta. PLoS ONE 2017, 12, e0170510. [Google Scholar]
- Harrison, R.L.; Mowery, J.D.; Rowley, D.L.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The complete genome sequence of a third distinct baculovirus isolated from the true armyworm, Mythimna unipuncta, contains two copies of the lef-7 gene. Virus Genes 2018, 54, 297–310. [Google Scholar] [CrossRef]
- Cuartas, P.; Barrera, G.; Belaich, M.; Barreto, E.; Ghiringhelli, P.; Villamizar, L. The complete sequence of the first Spodoptera frugiperda Betabaculovirus genome: A natural multiple recombinant virus. Viruses 2015, 7, 394–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrera, G.P.; Belaich, M.N.; Patarroyo, M.A.; Villamizar, L.F.; Ghiringhelli, P.D. Evidence of recent interspecies horizontal gene transfer regarding nucleopolyhedrovirus infection of Spodoptera frugiperda. BMC Genom. 2015, 16, 1008. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Yu, J.; Wang, L.; Hu, X.; Bao, W.; Li, G.; Chen, C.; Han, H.; Hu, S.; Yang, H. Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology 2001, 287, 391–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willis, L.G.; Siepp, R.; Stewart, T.M.; Erlandson, M.A.; Theilmann, D.A. Sequence analysis of the complete genome of Trichoplusia ni single nucleopolyhedrovirus and the identification of a baculoviral photolyase gene. Virology 2005, 338, 209–226. [Google Scholar] [CrossRef] [Green Version]
- de los Ángeles Bivian-Hernández, M.; López-Tlacomulco, J.; Mares-Mares, E.; Ibarra, J.E.; Del Rincón-Castro, M.C. Genomic analysis of a Trichoplusia ni Betabaculovirus (TnGV) with three different viral enhancing factors and two unique genes. Arch. Virol. 2017, 162, 3705–3715. [Google Scholar] [CrossRef] [PubMed]
- Kemp, E.M.; Woodward, D.T.; Cory, J.S. Detection of single and mixed covert baculovirus infections in eastern spruce budworm, Choristoneura fumiferana populations. J. Invertebr. Pathol. 2011, 107, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Hackett, K.J.; Boore, A.; Deming, C.; Buckley, E.; Camp, M.; Shapiro, M. Helicoverpa armigera granulovirus interference with progression of H. zea nucleopolyhedrovirus disease in H. zea larvae. J. Invertebr. Pathol. 2000, 75, 99–106. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Köhler, T.; Alletti, G.G.; Jehle, J.A. Mortality of cutworm larvae is not enhanced by agrotis segetum granulovirus and agrotis segetum nucleopolyhedrovirus b coinfection relative to single infection by either virus. Appl. Environ. Microbiol. 2015, 81, 2893–2899. [Google Scholar] [CrossRef] [Green Version]





| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SporGV | 0.013 | 0.011 | 0.013 | 0.012 | 0.012 | 0.012 | 0.012 | 0.012 | 0.010 | 0.009 | 0.010 | 0.012 | 0.012 | 0.010 | 0.011 | 0.011 | 0.009 | 0.004 | 0.010 | 0.010 | |
| 2 | AdorGV | 0.969 | 0.011 | 0.011 | 0.009 | 0.010 | 0.009 | 0.011 | 0.009 | 0.013 | 0.012 | 0.015 | 0.011 | 0.010 | 0.013 | 0.008 | 0.013 | 0.014 | 0.012 | 0.013 | 0.013 | |
| 3 | AgseGV | 0.844 | 0.873 | 0.012 | 0.011 | 0.012 | 0.010 | 0.012 | 0.011 | 0.011 | 0.011 | 0.013 | 0.012 | 0.012 | 0.011 | 0.010 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | |
| 4 | ClanGV | 0.961 | 0.812 | 0.937 | 0.009 | 0.008 | 0.009 | 0.011 | 0.009 | 0.013 | 0.014 | 0.013 | 0.009 | 0.011 | 0.014 | 0.008 | 0.013 | 0.013 | 0.013 | 0.014 | 0.013 | |
| 5 | CrleGV | 0.903 | 0.690 | 0.825 | 0.738 | 0.007 | 0.008 | 0.011 | 0.008 | 0.013 | 0.012 | 0.016 | 0.010 | 0.009 | 0.012 | 0.007 | 0.013 | 0.015 | 0.012 | 0.012 | 0.012 | |
| 6 | CypoGV | 0.890 | 0.786 | 0.875 | 0.675 | 0.492 | 0.009 | 0.010 | 0.008 | 0.012 | 0.013 | 0.011 | 0.008 | 0.009 | 0.012 | 0.007 | 0.011 | 0.011 | 0.013 | 0.012 | 0.012 | |
| 7 | DisaGV | 0.913 | 0.734 | 0.832 | 0.735 | 0.620 | 0.667 | 0.011 | 0.008 | 0.012 | 0.012 | 0.015 | 0.009 | 0.009 | 0.012 | 0.007 | 0.012 | 0.013 | 0.012 | 0.012 | 0.012 | |
| 8 | EpapGV | 0.921 | 0.863 | 0.869 | 0.863 | 0.835 | 0.781 | 0.833 | 0.011 | 0.012 | 0.012 | 0.012 | 0.010 | 0.011 | 0.013 | 0.010 | 0.011 | 0.012 | 0.012 | 0.013 | 0.012 | |
| 9 | ErelGV | 0.924 | 0.736 | 0.858 | 0.709 | 0.655 | 0.648 | 0.661 | 0.814 | 0.013 | 0.012 | 0.014 | 0.010 | 0.009 | 0.013 | 0.007 | 0.012 | 0.013 | 0.012 | 0.013 | 0.013 | |
| 10 | HearGV | 0.763 | 0.977 | 0.865 | 1.005 | 0.939 | 0.927 | 0.937 | 0.928 | 0.954 | 0.004 | 0.006 | 0.012 | 0.012 | 0.003 | 0.011 | 0.011 | 0.006 | 0.009 | 0.003 | 0.001 | |
| 11 | MolaGV | 0.750 | 0.937 | 0.868 | 1.015 | 0.902 | 0.943 | 0.924 | 0.934 | 0.927 | 0.339 | 0.006 | 0.013 | 0.012 | 0.005 | 0.011 | 0.011 | 0.006 | 0.009 | 0.005 | 0.004 | |
| 12 | MyunGV | 0.755 | 1.093 | 0.964 | 0.970 | 1.082 | 0.877 | 1.045 | 0.920 | 1.009 | 0.488 | 0.516 | 0.012 | 0.014 | 0.006 | 0.014 | 0.011 | 0.005 | 0.010 | 0.006 | 0.006 | |
| 13 | PlinGV | 0.897 | 0.812 | 0.876 | 0.731 | 0.731 | 0.663 | 0.699 | 0.770 | 0.724 | 0.933 | 0.950 | 0.876 | 0.010 | 0.012 | 0.009 | 0.011 | 0.012 | 0.012 | 0.012 | 0.012 | |
| 14 | PhopGV | 0.942 | 0.745 | 0.877 | 0.822 | 0.693 | 0.712 | 0.709 | 0.831 | 0.727 | 0.952 | 0.942 | 1.032 | 0.780 | 0.013 | 0.008 | 0.012 | 0.013 | 0.012 | 0.013 | 0.013 | |
| 15 | PsunGV | 0.764 | 0.985 | 0.865 | 1.008 | 0.922 | 0.939 | 0.926 | 0.942 | 0.962 | 0.223 | 0.333 | 0.485 | 0.936 | 0.960 | 0.011 | 0.011 | 0.006 | 0.009 | 0.000 | 0.003 | |
| 16 | PiraGV | 0.873 | 0.655 | 0.795 | 0.644 | 0.529 | 0.576 | 0.550 | 0.775 | 0.536 | 0.884 | 0.857 | 1.035 | 0.669 | 0.648 | 0.880 | 0.012 | 0.013 | 0.011 | 0.011 | 0.011 | |
| 17 | PlxyGV | 0.871 | 0.932 | 0.857 | 0.950 | 0.918 | 0.864 | 0.919 | 0.862 | 0.921 | 0.873 | 0.876 | 0.859 | 0.870 | 0.940 | 0.862 | 0.864 | 0.011 | 0.011 | 0.011 | 0.011 | |
| 18 | SpfrGV | 0.746 | 1.036 | 0.896 | 0.973 | 1.012 | 0.863 | 0.968 | 0.921 | 0.955 | 0.487 | 0.512 | 0.418 | 0.904 | 0.997 | 0.491 | 0.955 | 0.855 | 0.009 | 0.006 | 0.006 | |
| 19 | SpltGV | 0.309 | 0.935 | 0.823 | 0.991 | 0.877 | 0.928 | 0.891 | 0.918 | 0.918 | 0.739 | 0.729 | 0.783 | 0.928 | 0.923 | 0.741 | 0.848 | 0.858 | 0.761 | 0.009 | 0.009 | |
| 20 | TrniGV | 0.764 | 0.987 | 0.865 | 1.008 | 0.924 | 0.940 | 0.926 | 0.943 | 0.962 | 0.223 | 0.332 | 0.484 | 0.936 | 0.961 | 0.003 | 0.880 | 0.862 | 0.491 | 0.740 | 0.003 | |
| 21 | XecnGV | 0.771 | 0.983 | 0.867 | 1.004 | 0.937 | 0.925 | 0.938 | 0.933 | 0.952 | 0.030 | 0.339 | 0.486 | 0.935 | 0.958 | 0.221 | 0.886 | 0.876 | 0.487 | 0.743 | 0.221 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LeseNPV | 0.007 | 0.014 | 0.011 | 0.012 | 0.013 | 0.013 | 0.013 | 0.014 | 0.014 | 0.013 | 0.015 | 0.013 | 0.015 | 0.010 | 0.012 | 0.012 | 0.013 | 0.012 | 0.013 | 0.010 | 0.010 | 0.011 | 0.011 | 0.012 | 0.010 | 0.010 | |
| 2 | SpliNPV | 0.635 | 0.012 | 0.011 | 0.013 | 0.012 | 0.013 | 0.012 | 0.013 | 0.013 | 0.013 | 0.014 | 0.012 | 0.013 | 0.011 | 0.012 | 0.012 | 0.012 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | 0.011 | 0.012 | 0.011 | 0.011 | |
| 3 | ClbiNPV | 1.067 | 1.013 | 0.010 | 0.010 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.010 | 0.012 | 0.009 | 0.009 | 0.010 | 0.010 | 0.010 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | |
| 4 | HearNPV | 0.929 | 0.913 | 0.839 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.013 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.011 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | |
| 5 | LafiNPV | 0.983 | 1.003 | 0.825 | 0.895 | 0.009 | 0.009 | 0.008 | 0.009 | 0.009 | 0.009 | 0.009 | 0.008 | 0.011 | 0.009 | 0.011 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.011 | 0.010 | |
| 6 | PeluSNPV | 0.995 | 0.942 | 0.672 | 0.818 | 0.781 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.010 | 0.010 | 0.009 | 0.009 | 0.009 | 0.010 | 0.010 | 0.010 | 0.010 | 0.009 | 0.009 | 0.010 | 0.009 | 0.010 | |
| 7 | EcobNPV | 1.047 | 1.007 | 0.770 | 0.841 | 0.776 | 0.748 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.010 | 0.012 | 0.009 | 0.010 | 0.010 | 0.010 | 0.010 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | |
| 8 | EupsNPV | 1.009 | 0.984 | 0.756 | 0.853 | 0.726 | 0.734 | 0.665 | 0.008 | 0.008 | 0.007 | 0.008 | 0.007 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | |
| 9 | BusuNPV | 1.058 | 1.028 | 0.765 | 0.837 | 0.770 | 0.737 | 0.649 | 0.661 | 0.006 | 0.008 | 0.008 | 0.008 | 0.010 | 0.012 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.012 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.011 | |
| 10 | SujuNPV | 1.073 | 1.049 | 0.780 | 0.867 | 0.808 | 0.759 | 0.675 | 0.693 | 0.530 | 0.008 | 0.008 | 0.008 | 0.010 | 0.012 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.011 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.011 | |
| 11 | HespNPV | 1.022 | 1.008 | 0.797 | 0.861 | 0.789 | 0.741 | 0.665 | 0.642 | 0.673 | 0.685 | 0.008 | 0.007 | 0.010 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.011 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | |
| 12 | ApciNPV | 1.137 | 1.055 | 0.754 | 0.834 | 0.801 | 0.728 | 0.652 | 0.674 | 0.643 | 0.665 | 0.665 | 0.008 | 0.010 | 0.014 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.013 | 0.012 | 0.010 | 0.010 | 0.011 | 0.011 | 0.012 | |
| 13 | OrleNPV | 0.994 | 0.994 | 0.760 | 0.844 | 0.715 | 0.732 | 0.654 | 0.591 | 0.632 | 0.665 | 0.620 | 0.657 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.009 | 0.010 | |
| 14 | AdhoNPV | 1.096 | 1.042 | 0.836 | 0.862 | 0.906 | 0.808 | 0.840 | 0.840 | 0.830 | 0.844 | 0.853 | 0.804 | 0.833 | 0.014 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.013 | 0.012 | 0.010 | 0.010 | 0.010 | 0.011 | 0.011 | |
| 15 | LydiMNPV | 0.860 | 0.953 | 0.896 | 0.992 | 0.808 | 0.829 | 0.919 | 0.832 | 0.915 | 0.932 | 0.874 | 1.043 | 0.820 | 1.052 | 0.012 | 0.012 | 0.012 | 0.012 | 0.012 | 0.008 | 0.008 | 0.011 | 0.011 | 0.011 | 0.010 | 0.009 | |
| 16 | TrniSNPV | 0.977 | 0.965 | 0.793 | 0.824 | 0.877 | 0.767 | 0.806 | 0.825 | 0.804 | 0.825 | 0.811 | 0.794 | 0.803 | 0.834 | 0.942 | 0.002 | 0.002 | 0.008 | 0.008 | 0.009 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| 17 | ChchNPV | 0.967 | 0.960 | 0.798 | 0.827 | 0.879 | 0.769 | 0.807 | 0.828 | 0.808 | 0.845 | 0.830 | 0.802 | 0.815 | 0.840 | 0.927 | 0.164 | 0.002 | 0.008 | 0.008 | 0.009 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| 18 | PsinSNPV | 0.977 | 0.976 | 0.807 | 0.826 | 0.876 | 0.774 | 0.817 | 0.827 | 0.806 | 0.848 | 0.826 | 0.799 | 0.806 | 0.842 | 0.931 | 0.176 | 0.167 | 0.009 | 0.008 | 0.009 | 0.008 | 0.009 | 0.009 | 0.009 | 0.008 | 0.008 | |
| 19 | HearMNPV | 0.986 | 0.972 | 0.825 | 0.816 | 0.857 | 0.796 | 0.820 | 0.818 | 0.811 | 0.843 | 0.843 | 0.820 | 0.815 | 0.863 | 0.923 | 0.699 | 0.695 | 0.706 | 0.000 | 0.007 | 0.006 | 0.006 | 0.006 | 0.007 | 0.007 | 0.006 | |
| 20 | MabrMNPV | 0.990 | 0.976 | 0.829 | 0.819 | 0.861 | 0.799 | 0.823 | 0.822 | 0.814 | 0.845 | 0.844 | 0.823 | 0.817 | 0.863 | 0.925 | 0.700 | 0.697 | 0.707 | 0.008 | 0.007 | 0.006 | 0.006 | 0.007 | 0.007 | 0.007 | 0.006 | |
| 21 | PespNPV | 0.819 | 0.911 | 0.879 | 0.907 | 0.825 | 0.832 | 0.893 | 0.833 | 0.888 | 0.919 | 0.892 | 0.989 | 0.827 | 0.982 | 0.669 | 0.749 | 0.750 | 0.748 | 0.549 | 0.553 | 0.005 | 0.006 | 0.006 | 0.006 | 0.006 | 0.005 | |
| 22 | AgipMNPV | 0.844 | 0.908 | 0.841 | 0.857 | 0.829 | 0.807 | 0.861 | 0.818 | 0.871 | 0.892 | 0.843 | 0.908 | 0.806 | 0.927 | 0.725 | 0.705 | 0.703 | 0.711 | 0.527 | 0.530 | 0.401 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | |
| 23 | SfMNPV 3AP2 | 0.933 | 0.928 | 0.830 | 0.824 | 0.839 | 0.794 | 0.814 | 0.816 | 0.822 | 0.840 | 0.831 | 0.830 | 0.813 | 0.874 | 0.883 | 0.706 | 0.703 | 0.717 | 0.558 | 0.562 | 0.552 | 0.448 | 0.000 | 0.001 | 0.004 | 0.004 | |
| 24 | SfMNPV Col | 0.933 | 0.928 | 0.829 | 0.825 | 0.839 | 0.794 | 0.814 | 0.816 | 0.821 | 0.839 | 0.829 | 0.829 | 0.814 | 0.873 | 0.883 | 0.707 | 0.703 | 0.718 | 0.558 | 0.562 | 0.552 | 0.447 | 0.005 | 0.001 | 0.004 | 0.004 | |
| 25 | SporNPV | 0.931 | 0.933 | 0.831 | 0.830 | 0.841 | 0.801 | 0.819 | 0.815 | 0.827 | 0.840 | 0.831 | 0.837 | 0.813 | 0.877 | 0.873 | 0.712 | 0.705 | 0.718 | 0.560 | 0.564 | 0.548 | 0.441 | 0.025 | 0.021 | 0.004 | 0.004 | |
| 26 | SpexMNPV | 0.906 | 0.902 | 0.838 | 0.835 | 0.859 | 0.798 | 0.825 | 0.816 | 0.837 | 0.865 | 0.831 | 0.864 | 0.807 | 0.890 | 0.810 | 0.699 | 0.704 | 0.705 | 0.560 | 0.556 | 0.503 | 0.407 | 0.360 | 0.360 | 0.343 | 0.003 | |
| 27 | SpltMNPV | 0.869 | 0.902 | 0.831 | 0.842 | 0.838 | 0.806 | 0.832 | 0.812 | 0.852 | 0.880 | 0.829 | 0.894 | 0.811 | 0.901 | 0.767 | 0.704 | 0.705 | 0.706 | 0.559 | 0.562 | 0.472 | 0.384 | 0.343 | 0.342 | 0.323 | 0.205 |
| Insect | Slope (±SE) | LC50 (OBs/mL) | Fiducial Limits (95%) | df | χ2 | |
|---|---|---|---|---|---|---|
| Low | High | |||||
| S. ornithogalli | 1.29 (0.15) | 4.87 × 104 | 2.88 × 104 | 8.24 × 104 | 3 | 5.22 |
| Treatment | Slope (±SE) | LC50 (OBs/mL) | Relative Potency | Fiducial Limits (95%) | p Value | |
| Low | High | |||||
| SporNPV | 1.03 (0.11) | 1.66 × 105 a | 1.04 × 105 | 2.60 × 105 | ||
| M1 | 1.04 (0.12) | 1.23 × 105 ab | 1.35 | 7.17 × 104 | 2.11 × 105 | 0.05 |
| M2 | 1.38 (0.16) | 7.39 × 104 ab | 2.25 | 4.22 × 104 | 1.28 × 105 | 0.45 |
| M3 | 1.38 (0.18) | 5.42 × 104 b | 3.06 | 3.03 × 104 | 9.48 × 104 | <0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrera, G.P.; Villamizar, L.F.; Araque, G.A.; Gómez, J.A.; Guevara, E.J.; Cerrudo, C.S.; Belaich, M.N. Natural Coinfection between Novel Species of Baculoviruses in Spodoptera ornithogalli Larvae. Viruses 2021, 13, 2520. https://doi.org/10.3390/v13122520
Barrera GP, Villamizar LF, Araque GA, Gómez JA, Guevara EJ, Cerrudo CS, Belaich MN. Natural Coinfection between Novel Species of Baculoviruses in Spodoptera ornithogalli Larvae. Viruses. 2021; 13(12):2520. https://doi.org/10.3390/v13122520
Chicago/Turabian StyleBarrera, Gloria Patricia, Laura Fernanda Villamizar, Gustavo Adolfo Araque, Juliana Andrea Gómez, Elsa Judith Guevara, Carolina Susana Cerrudo, and Mariano Nicolás Belaich. 2021. "Natural Coinfection between Novel Species of Baculoviruses in Spodoptera ornithogalli Larvae" Viruses 13, no. 12: 2520. https://doi.org/10.3390/v13122520
APA StyleBarrera, G. P., Villamizar, L. F., Araque, G. A., Gómez, J. A., Guevara, E. J., Cerrudo, C. S., & Belaich, M. N. (2021). Natural Coinfection between Novel Species of Baculoviruses in Spodoptera ornithogalli Larvae. Viruses, 13(12), 2520. https://doi.org/10.3390/v13122520