SARS CoV-2 (Delta Variant) Infection Kinetics and Immunopathogenesis in Domestic Cats
Abstract
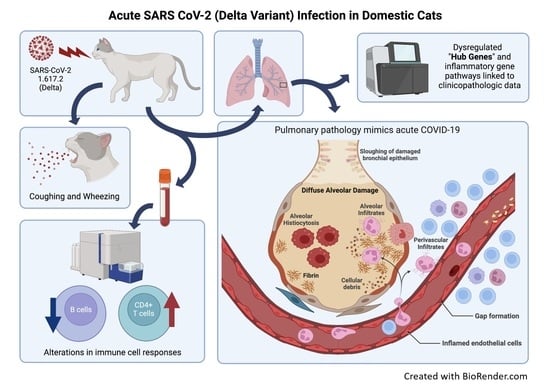
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Animals
2.3. Virus Challenge
2.4. Clinical Evaluation
2.5. Sample Collection
2.6. Virus Isolation
2.7. Histopathology
2.8. Immunofluorescence Assay (IFA)
2.9. Flow Cytometry
2.10. Viral Genome Sequencing, Genome Assembly, Alignment and Phylogenetic Analysis
2.11. Viral RNA Analysis
2.12. Feline ACE-2 Analysis
2.13. Lung Transcriptome Analysis
2.14. Statistical Analysis
3. Results
3.1. SARS-CoV-2 (Delta Variant)-Infected Cats Develop Acute Clinical Respiratory Disease
3.2. Infectious Virus Is Shed via Nasal Secretions up to 4 Days Post-Inoculation
3.3. Sars-Cov-2 (B.1.617.2) Causes Severe Lung Pathology in Domestic Cats Akin to COVID-19
3.4. Sars-Cov-2 Antigen Is Detected in ACE2-Expressing Lungs of Domestic Cats during Infection with the Delta Variant
3.5. Domestic Cats Exhibit Divergent Lymphocyte Immunophenotypes during SARS-CoV-2 Infection
3.6. Viral Sequencing and Genome Assembly
3.7. Viral RNA and ACE-2 Expression in Tissues during SARS CoV-2 (Delta Variant) Infection
3.8. Inflammatory Pathways Are Dysregulated in Acute Phase and Neurological Pathways Are Altered in Chronic Phase with a Shift in Transcriptomic Profile from One Phase to Another
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Li, B.; Deng, A.; Li, K.; Hu, Y.; Li, Z.; Shi, Y.; Xiong, Q.; Liu, Z.; Guo, Q.; Zou, L. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat. Commun. 2022, 13, 460. [Google Scholar] [CrossRef]
- Sheikh, A.; McMenamin, J.; Taylor, B.; Robertson, C. SARS-CoV-2 Delta VOC in Scotland: Demographics, risk of hospital admission, and vaccine effectiveness. Lancet 2021, 397, 2461–2462. [Google Scholar] [CrossRef]
- Fisman, D.N.; Tuite, A.R. Evaluation of the relative virulence of novel SARS-CoV-2 variants: A retrospective cohort study in Ontario, Canada. Cmaj 2021, 193, E1619–E1625. [Google Scholar] [CrossRef] [PubMed]
- Pung, R.; Mak, T.M.; Kucharski, A.J.; Lee, V.J. Serial intervals in SARS-CoV-2 B. 1.617. 2 variant cases. Lancet 2021, 398, 837–838. [Google Scholar] [CrossRef]
- Zhang, M.; Xiao, J.; Deng, A.; Zhang, Y.; Zhuang, Y.; Hu, T.; Li, J.; Tu, H.; Li, B.; Zhou, Y. Transmission dynamics of an outbreak of the COVID-19 Delta variant B. 1.617. 2—Guangdong Province, China, May–June 2021. China CDC Wkly. 2021, 3, 584. [Google Scholar] [CrossRef] [PubMed]
- GISAID. Number of SARS-CoV-2 Delta Variant Cases Worldwide as of 19 April 2022, by Country or Territory. 2022. Available online: https://www.gisaid.org/hcov19-variants/ (accessed on 28 April 2022).
- Wang, Y.; Chen, R.; Hu, F.; Lan, Y.; Yang, Z.; Zhan, C.; Shi, J.; Deng, X.; Jiang, M.; Zhong, S. Transmission, viral kinetics and clinical characteristics of the emergent SARS-CoV-2 Delta VOC in Guangzhou, China. EClinicalMedicine 2021, 40, 101129. [Google Scholar] [CrossRef]
- Chenchula, S.; Karunakaran, P. Current Evidence on Efficacy of COVID-19 Booster Dose Vaccination Against the Omicron Variant. A Systematic Review. J. Med. Virol. 2022, 94, 2969–2976. [Google Scholar] [CrossRef]
- Gardner, B.J.; Kilpatrick, A.M. Estimates of reduced vaccine effectiveness against hospitalization, infection, transmission and symptomatic disease of a new SARS-CoV-2 variant, Omicron (B. 1.1. 529), using neutralizing antibody titers. MedRxiv. [CrossRef]
- Callaway, E. Omicron likely to weaken COVID vaccine protection. Nature 2021, 600, 367–368. [Google Scholar] [CrossRef]
- Cool, K.; Gaudreault, N.N.; Morozov, I.; Trujillo, J.D.; Meekins, D.A.; McDowell, C.; Carossino, M.; Bold, D.; Mitzel, D.; Kwon, T. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg. Microbes Infect. 2022, 11, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Gunasekara, S.; Tamil Selvan, M.; Miller, C.A.; Rudd, J.M. Thinking Outside the Box: Utilizing Nontraditional Animal Models for COVID-19 Research. Int. J. Transl. Med. 2022, 2, 113–133. [Google Scholar] [CrossRef]
- Kim, Y.I.; Casel, M.A.B.; Choi, Y.K. Transmissibility and pathogenicity of SARS-CoV-2 variants in animal models. J. Microbiol. 2022, 60, 255–267. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Lowen, A.C. Animal models for SARS-CoV-2. Curr. Opin. Virol. 2021, 48, 73–81. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Widerspick, L.; Albrecht, R.A.; Beer, M.; Carroll, M.W.; de Wit, E.; Diamond, M.S.; Dowling, W.E.; Funnell, S.G.; García-Sastre, A. Advances and gaps in SARS-CoV-2 infection models. PLoS Pathog. 2022, 18, e1010161. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.-D.; Liu, M.-Q.; Chen, Y.; Shan, C.; Zhou, Y.-W.; Shen, X.-R.; Li, Q.; Zhang, L.; Zhu, Y.; Si, H.-R. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 2020, 182, 50–58.e8. [Google Scholar] [CrossRef]
- Ryan, K.A.; Bewley, K.R.; Fotheringham, S.A.; Slack, G.S.; Brown, P.; Hall, Y.; Wand, N.I.; Marriott, A.C.; Cavell, B.E.; Tree, J.A. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat. Commun. 2021, 12, 81. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.-M.; Chin, A.W.; Fung, K.; Choy, K.-T.; Wong, A.Y.; Kaewpreedee, P.; Perera, R.A.; Poon, L.L.; Nicholls, J.M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zhao, Y.; Yu, W.; Yang, Y.; Gao, J.; Wang, J.; Kuang, D.; Yang, M.; Yang, J.; Ma, C. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct. Target. Ther. 2020, 5, 1–9. [Google Scholar] [CrossRef]
- Shuai, L.; Zhong, G.; Yuan, Q.; Wen, Z.; Wang, C.; He, X.; Liu, R.; Wang, J.; Zhao, Q.; Liu, Y. Replication, pathogenicity, and transmission of SARS-CoV-2 in minks. Natl. Sci. Rev. 2021, 8, nwaa291. [Google Scholar] [CrossRef] [PubMed]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.-I.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020, 383, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Hosie, M.J.; Hofmann-Lehmann, R.; Hartmann, K.; Egberink, H.; Truyen, U.; Addie, D.D.; Belák, S.; Boucraut-Baralon, C.; Frymus, T.; Lloret, A. Anthropogenic infection of cats during the 2020 COVID-19 pandemic. Viruses 2021, 13, 185. [Google Scholar] [CrossRef]
- Rudd, J.M.; Tamil Selvan, M.; Cowan, S.; Kao, Y.-F.; Midkiff, C.C.; Narayanan, S.; Ramachandran, A.; Ritchey, J.W.; Miller, C.A. Clinical and Histopathologic Features of a Feline SARS-CoV-2 Infection Model Are Analogous to Acute COVID-19 in Humans. Viruses 2021, 13, 1550. [Google Scholar] [CrossRef]
- Bao, L.; Song, Z.; Xue, J.; Gao, H.; Liu, J.; Wang, J.; Guo, Q.; Zhao, B.; Qu, Y.; Qi, F.; et al. Susceptibility and Attenuated Transmissibility of SARS-CoV-2 in Domestic Cats. J. Infect. Dis. 2021, 223, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Carossino, M.; Morozov, I.; Trujillo, J.D.; Meekins, D.A.; Madden, D.W.; Cool, K.; Artiaga, B.L.; McDowell, C.; Bold, D. Experimental re-infected cats do not transmit SARS-CoV-2. Emerging Microbes & Infections 2021, 10, 638–650. [Google Scholar] [CrossRef]
- Hosie, M.J.; Epifano, I.; Herder, V.; Orton, R.J.; Stevenson, A.; Johnson, N.; MacDonald, E.; Dunbar, D.; McDonald, M.; Howie, F.; et al. Detection of SARS-CoV-2 in respiratory samples from cats in the UK associated with human-to-cat transmission. Vet. Rec. 2021, 188, e247. [Google Scholar] [CrossRef]
- Kang, K.; Chen, Q.; Gao, Y.; Yu, K.J. Detection of SARS-CoV-2 B. 1.617. 2 (Delta) variant in three cats owned by a confirmed COVID-19 patient in Harbin, China. Vet. Med. Sci. 2021, 8, 945. [Google Scholar] [CrossRef]
- Karikalan, M.; Chander, V.; Mahajan, S.; Deol, P.; Agrawal, R.; Nandi, S.; Rai, S.; Mathur, A.; Pawde, A.; Singh, K. Natural infection of Delta mutant of SARS-CoV-2 in Asiatic lions of India. Transbound. Emerg. Dis. 2021, 1–9. [Google Scholar] [CrossRef]
- Lenz, O.C.; Marques, A.D.; Kelly, B.J.; Rodino, K.G.; Cole, S.D.; Perera, R.A.; Weiss, S.R.; Bushman, F.D.; Lennon, E.M. SARS-CoV-2 Delta Variant (AY. 3) in the Feces of a Domestic Cat. Viruses 2022, 14, 421. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Kumar, N.; Bhatia, S.; Aasdev, A.; Kanniappan, S.; Sekhar, A.T.; Gopinadhan, A.; Silambarasan, R.; Sreekumar, C.; Dubey, C.K. Sars-cov-2 delta variant among asiatic lions, india. Emerg. Infect. Dis. 2021, 27, 2723. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H.J.A.j.o.e. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Ritchey, J.W.; Levy, J.K.; Bliss, S.K.; Tompkins, W.A.; Tompkins, M.B. Constitutive expression of types 1 and 2 cytokines by alveolar macrophages from feline immunodeficiency virus-infected cats. Vet. Immunol. Immunopathol. 2001, 79, 83–100. [Google Scholar] [CrossRef]
- Miller, C.; Bielefeldt-Ohmann, H.; MacMillan, M.; Huitron-Resendiz, S.; Henriksen, S.; Elder, J.; VandeWoude, S. Strain-specific viral distribution and neuropathology of feline immunodeficiency virus. Vet. Immunol. Immunopathol. 2011, 143, 282–291. [Google Scholar] [CrossRef] [Green Version]
- Polak, S.B.; Van Gool, I.C.; Cohen, D.; Jan, H.; van Paassen, J. A systematic review of pathological findings in COVID-19: A pathophysiological timeline and possible mechanisms of disease progression. Mod. Pathol. 2020, 33, 2128–2138. [Google Scholar] [CrossRef]
- Tian, S.; Xiong, Y.; Liu, H.; Niu, L.; Guo, J.; Liao, M.; Xiao, S.-Y. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod. Pathol. 2020, 33, 1007–1014. [Google Scholar] [CrossRef] [Green Version]
- von der Thüsen, J.; van der Eerden, M. Histopathology and genetic susceptibility in COVID-19 pneumonia. Eur. J. Clin. Investig. 2020, 50, e13259. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Miller, C.; Boegler, K.; Carver, S.; MacMillan, M.; Bielefeldt-Ohmann, H.; VandeWoude, S. Pathogenesis of oral FIV infection. PLoS ONE 2017, 12, e0185138. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef]
- Pickett, B.E.; Sadat, E.L.; Zhang, Y.; Noronha, J.M.; Squires, R.B.; Hunt, V.; Liu, M.; Kumar, S.; Zaremba, S.; Gu, Z. ViPR: An open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012, 40, D593–D598. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA, X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Powers, J.; Musselman, E.; Mackie, R.; Elder, J.; VandeWoude, S. Immunopathologic Effects of Prednisolone and Cyclosporine A on Feline Immunodeficiency Virus Replication and Persistence. Viruses 2019, 11, 805. [Google Scholar] [CrossRef] [Green Version]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Buckley, R.M.; Davis, B.W.; Brashear, W.A.; Farias, F.H.G.; Kuroki, K.; Graves, T.; Hillier, L.W.; Kremitzki, M.; Li, G.; Middleton, R.P.; et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. PLoS Genet 2020, 16, e1008926. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, H.L.; Limtung, P. Mutations in the phosphorylation sites of SARS-CoV-2 encoded nucleocapsid protein and structure model of sequestration by protein 14-3-3. Biochem. Biophys. Res. Commun. 2020, 532, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Prüβ, B.M. Variants of SARS CoV-2: Mutations, transmissibility, virulence, drug resistance, and antibody/vaccine sensitivity. Front. Biosci.-Landmark 2022, 27, 65. [Google Scholar] [CrossRef] [PubMed]
- Barroso-Arévalo, S.; Sánchez-Morales, L.; Pérez-Sancho, M.; Domínguez, L.; Sánchez-Vizcaíno, J.M. First detection of SARS-CoV-2 B. 1.617. 2 (Delta) variant of concern in a symptomatic cat in Spain. Front. Vet. Sci. 2022, 9, 841430. [Google Scholar] [CrossRef] [PubMed]
- Essa, R.A.; Ahmed, S.K.; Bapir, D.H.; Rasul, S.A.; Khdir, A.A.; Abubakr, C.P. Clinical features and laboratory findings first case of B. 1.617. 2 (delta) variant concern (VOC) in Iraq. Ann. Med. Surg. 2021, 69, 102814. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Singanayagam, A.; Hakki, S.; Dunning, J.; Madon, K.J.; Crone, M.A.; Koycheva, A.; Derqui-Fernandez, N.; Barnett, J.L.; Whitfield, M.G.; Varro, R. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B. 1.617. 2) variant in vaccinated and unvaccinated individuals in the UK: A prospective, longitudinal, cohort study. Lancet Infect. Dis. 2022, 22, 183–195. [Google Scholar] [CrossRef]
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: Estimation and application. Ann. Intern. Med. 2020, 172, 577–582. [Google Scholar] [CrossRef] [Green Version]
- Chaudhry, F.; Lavandero, S.; Xie, X.; Sabharwal, B.; Zheng, Y.-Y.; Correa, A.; Narula, J.; Levy, P. Manipulation of ACE2 expression in COVID-19. Open Heart 2020, 7, e001424. [Google Scholar] [CrossRef]
- Riou, C.; Du Bruyn, E.; Stek, C.; Daroowala, R.; Goliath, R.T.; Abrahams, F.; Said-Hartley, Q.; Allwood, B.W.; Hsiao, N.-Y.; Wilkinson, K.A. Relationship of SARS-CoV-2–specific CD4 response to COVID-19 severity and impact of HIV-1 and tuberculosis coinfection. J. Clin. Investig. 2021, 131, 12. [Google Scholar] [CrossRef] [PubMed]
- Vardhana, S.; Baldo, L.; Morice, W.G.; Wherry, E.J. Understanding T-cell responses to COVID-19 is essential for informing public health strategies. Sci. Immunol. 2022, 7, eabo1303. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Hernández, V.A.; Torres-Ruíz, J.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Páez-Franco, J.C.; Meza-Sánchez, D.E.; Juárez-Vega, G.; Pérez-Fragoso, A.; Ortiz-Navarrete, V.; Ponce-de-León, A. B cell subsets as severity-associated signatures in COVID-19 patients. Front. Immunol. 2020, 3244. [Google Scholar] [CrossRef] [PubMed]
- De Biasi, S.; Lo Tartaro, D.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Mattioli, M.; Paolini, A.; Gozzi, L. Expansion of plasmablasts and loss of memory B cells in peripheral blood from COVID-19 patients with pneumonia. Eur. J. Immunol. 2020, 50, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.E.; Varga, S.M. The CD8 T cell response to respiratory virus infections. Front. Immunol. 2018, 678. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-I.; Kang, J.-H.; Kim, E.-H.; Seo, Y.-J. KIF11 inhibition decreases cytopathogenesis and replication of influenza A virus. Mol. Cell. Toxicol. 2021, 17, 201–212. [Google Scholar] [CrossRef]
- Velavan, T.P.; Pallerla, S.R.; Rüter, J.; Augustin, Y.; Kremsner, P.G.; Krishna, S.; Meyer, C.G. Host genetic factors determining COVID-19 susceptibility and severity. EBioMedicine 2021, 72, 103629. [Google Scholar] [CrossRef]
- Mamoor, S. BATF2 and FOXJ1 are differentially expressed in coronavirus infections. 2020. Available online: https://osf.io/psd97/download (accessed on 25 April 2022).
- Pawlica, P.; Yario, T.A.; White, S.; Wang, J.; Moss, W.N.; Hui, P.; Vinetz, J.M.; Steitz, J.A. SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2116668118. [Google Scholar] [CrossRef]
- Auwul, M.R.; Rahman, M.R.; Gov, E.; Shahjaman, M.; Moni, M.A. Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19. Brief. Bioinform. 2021, 22, bbab120. [Google Scholar] [CrossRef]
- Farahani, M.; Niknam, Z.; Amirabad, L.M.; Amiri-Dashatan, N.; Koushki, M.; Nemati, M.; Pouya, F.D.; Rezaei-Tavirani, M.; Rasmi, Y.; Tayebi, L. Molecular pathways involved in COVID-19 and potential pathway-based therapeutic targets. Biomed. Pharmacother. 2022, 145, 112420. [Google Scholar] [CrossRef]
- Azad, G.K. Identification and molecular characterization of mutations in nucleocapsid phosphoprotein of SARS-CoV-2. PeerJ 2021, 9, e10666. [Google Scholar] [CrossRef] [PubMed]
- Julià, A.; Bonafonte-Pardàs, I.; Gómez, A.; López-Lasanta, M.; López-Corbeto, M.; Martínez-Mateu, S.H.; Lladós, J.; Rodríguez-Nunez, I.; Myers, R.M.; Marsal, S. Targeting of the CD80/86 proinflammatory axis as a therapeutic strategy to prevent severe COVID-19. Sci. Rep. 2021, 11, 11462. [Google Scholar] [CrossRef] [PubMed]
- Peach, R.J.; Bajorath, J.; Naemura, J.; Leytze, G.; Greene, J.; Aruffo, A.; Linsley, P.S. Both Extracellular Immunoglobin-like Domains of CD80 Contain Residues Critical for Binding T Cell Surface Receptors CTLA-4 and CD28 (∗). J. Biol. Chem. 1995, 270, 21181–21187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.J.; Guevara, M.G.; Lin, E.; O’Leary, S.E. Single-Molecule Dynamics of SARS-CoV-2 5′Cap Recognition by Human eIF4F. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ercan, H.; Schrottmaier, W.C.; Pirabe, A.; Schmuckenschlager, A.; Pereyra, D.; Santol, J.; Pawelka, E.; Traugott, M.T.; Schörgenhofer, C.; Seitz, T. Platelet phenotype analysis of COVID-19 patients reveals progressive changes in the activation of integrin αIIbβ3, F13A1, the SARS-CoV-2 Target EIF4A1 and Annexin A5. Front. Cardiovasc. Med. 2021, 8, 779073. [Google Scholar] [CrossRef]








| Clinical Parameter | 0 (Healthy) | 1 | 2 | 3 |
|---|---|---|---|---|
| Body Weight | No weight loss | 0 to 5% weight loss | 5 to 10% weight loss | >10% weight loss |
| Temperature | 37.2 to 39.2 °C | 39.1 to 39.4 °C | 39.5 to 39.7 °C | >39.7 °C |
| SpO2 | 98 to 100% | 95 to 97% | 93 to 94% | <93% |
| Activity | Normal | Mild reduction when disturbed * (mild lethargy) | Moderate reduction when disturbed * (moderate lethargy) | Little to no activity disturbed * and reduced activity when stimulated ** |
| Behavior | Normal | Reduced interest in food and/or attention | Markedly reduced interest in food and/or attention | Anorexia and/or complete lack of interest |
| Respiratory Effort | Normal resting respiratory rate and normal effort | Mild tachypnea (>35 breaths per minute); no overt increase in effort | Moderate tachypnea (>40 breaths per minute); moderate increase in effort | Marked tachypnea (>45 breaths per minute); marked effort or dyspnea |
| Ocular and/or Nasal Discharge | None | Mild discharge noted | Moderate discharge noted | Marked discharge noted |
| Coughing | None | Occasional, rare cough | Intermittent coughing (at least one episode in 30 min) | Marked persistent coughing(2 or more episodes over 30 min) |
| Wheezing | None | Occasional, rare wheezing | Intermittent wheezing | Marked, frequent wheezing |
| ISOLATE ID | Inoculation Days | Total Number of Reads | Genome Coverage | % Draft Genome Assembled |
|---|---|---|---|---|
| Viral RNA | Day 0 | 724,000 | 7264.99× | 97.71% |
| 9671 Cr. Lung | Day 4 | 660,000 | 6622.52× | 95.5% |
| 9671 Tbln | Day 4 | 524,000 | 5257.88× | 96.55% |
| Nt Position | Genomic Region | hCoV-19/USA/PHC658/2021 | Viral RNA | Lung | TBLN | Mutation Type |
|---|---|---|---|---|---|---|
| 13131 | ORF 1ab | A | A | G | A | Synonymous |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamil Selvan, M.; Gunasekara, S.; Xiao, P.; Griffin, K.; Cowan, S.R.; Narayanan, S.; Ramachandran, A.; Hagen, D.E.; Ritchey, J.W.; Rudd, J.M.; et al. SARS CoV-2 (Delta Variant) Infection Kinetics and Immunopathogenesis in Domestic Cats. Viruses 2022, 14, 1207. https://doi.org/10.3390/v14061207
Tamil Selvan M, Gunasekara S, Xiao P, Griffin K, Cowan SR, Narayanan S, Ramachandran A, Hagen DE, Ritchey JW, Rudd JM, et al. SARS CoV-2 (Delta Variant) Infection Kinetics and Immunopathogenesis in Domestic Cats. Viruses. 2022; 14(6):1207. https://doi.org/10.3390/v14061207
Chicago/Turabian StyleTamil Selvan, Miruthula, Sachithra Gunasekara, Ping Xiao, Kristen Griffin, Shannon R. Cowan, Sai Narayanan, Akhilesh Ramachandran, Darren E. Hagen, Jerry W. Ritchey, Jennifer M. Rudd, and et al. 2022. "SARS CoV-2 (Delta Variant) Infection Kinetics and Immunopathogenesis in Domestic Cats" Viruses 14, no. 6: 1207. https://doi.org/10.3390/v14061207
APA StyleTamil Selvan, M., Gunasekara, S., Xiao, P., Griffin, K., Cowan, S. R., Narayanan, S., Ramachandran, A., Hagen, D. E., Ritchey, J. W., Rudd, J. M., & Miller, C. A. (2022). SARS CoV-2 (Delta Variant) Infection Kinetics and Immunopathogenesis in Domestic Cats. Viruses, 14(6), 1207. https://doi.org/10.3390/v14061207