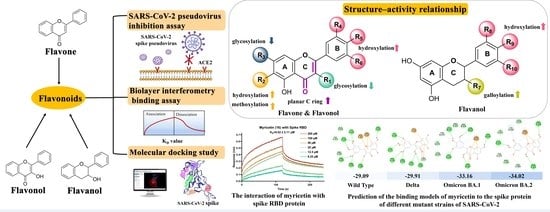
Anti-Entry Activity of Natural Flavonoids against SARS-CoV-2 by Targeting Spike RBD
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Cell Culture
2.3. Cytotoxicity Assay
2.4. Evaluation of SARS-CoV-2 Spike Pseudotyped Virus Entry into ACE2h Cells
2.5. Biolayer Interferometry (BLI) Binding Assay
2.6. Molecular Docking Study
2.7. Statistical Analysis
3. Results and Discussion
3.1. Inhibitory Activities of Flavonoids against the Entry of SARS-CoV-2 Pseudovirus into Host Cells and Structure–Activity Relationship Analysis
3.2. Binding Affinities of Selected Antiviral Entry Flavonoids with SARS-CoV-2 Spike RBD Protein
3.3. Identification of Potential Binding Pockets and Binding Mode for Bioactive Flavonoids Targeting SARS-CoV-2 Spike RBD
3.4. Cytotoxic Evaluation of Flavonoids in Human Normal Cell Lines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kesheh, M.M.; Hosseini, P.; Soltani, S.; Zandi, M. An overview on the seven pathogenic human coronaviruses. Rev. Med. Virol. 2022, 32, e2282. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Khandia, R.; Singhal, S.; Alqahtani, T.; Kamal, M.A.; Nahed, A.; Nainu, F.; Desingu, P.A.; Dhama, K. Emergence of SARS-CoV-2 Omicron (B. 1.1. 529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res. 2022, 209, 112816. [Google Scholar] [CrossRef]
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). 2022. Available online: https://coronavirus.jhu.edu/map.html (accessed on 5 December 2022).
- Grauer, J.; Löwen, H.; Liebchen, B. Strategic spatiotemporal vaccine distribution increases the survival rate in an infectious disease like COVID-19. Sci. Rep. 2020, 10, 21594. [Google Scholar] [CrossRef]
- Schwarzendahl, F.J.; Grauer, J.; Liebchen, B.; Löwen, H. Mutation induced infection waves in diseases like COVID-19. Sci. Rep. 2022, 12, 9641. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, C.; Ye, C.; Ruan, Z.; Liang, Y.; Li, Y.; Wu, J.; Luo, Z. Advances in the development of therapeutic strategies against COVID-19 and perspectives in the drug design for emerging SARS-CoV-2 variants. Comput. Struct. Biotechnol. J. 2022, 20, 824–837. [Google Scholar] [CrossRef]
- Thomas, E.; Stewart, L.E.; Darley, B.A.; Pham, A.M.; Esteban, I.; Panda, S.S. Plant-Based Natural Products and Extracts: Potential Source to Develop New Antiviral Drug Candidates. Molecules 2021, 26, 6197. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Alam, M.; Khatoon, F.; Fatima, U.; Elasbali, A.M.; Adnan, M.; Islam, A.; Hassan, M.I.; Snoussi, M.; De Feo, V. Natural products can be used in therapeutic management of COVID-19: Probable mechanistic insights. Biomed. Pharmacother. 2022, 147, 112658. [Google Scholar] [CrossRef]
- Martin, K.W.; Ernst, E. Antiviral agents from plants and herbs: A systematic review. Antivir. Ther. 2003, 8, 77–90. [Google Scholar] [CrossRef]
- Kumar, B.K.; Faheem, N.; Sekhar, K.V.G.C.; Ojha, R.; Prajapati, V.K.; Pai, A.; Murugesan, S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J. Biomol. Struct. Dyn. 2022, 40, 1363–1386. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Y.; Yao, S.; Ge, H.; Zhu, Y.; Chen, K.; Chen, W.-Z.; Zhang, Y.; Zhu, W.; Wang, H.-Y. Discovery of potential small molecular SARS-CoV-2 entry blockers targeting the spike protein. Acta Pharmacol. Sin. 2022, 43, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Twilley, D.; Esmear, T.; Oosthuizen, C.B.; Reid, A.-M.; Nel, M.; Lall, N. Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19). Front. Pharmacol. 2020, 11, 1514. [Google Scholar] [CrossRef] [PubMed]
- Badshah, S.L.; Faisal, S.; Muhammad, A.; Poulson, B.G.; Emwas, A.H.; Jaremko, M. Antiviral activities of flavonoids. Biomed. Pharmacother. 2021, 140, 111596. [Google Scholar] [CrossRef]
- Wang, T.-Y.; Li, Q.; Bi, K.-S. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef]
- Ahmad, A.; Kaleem, M.; Ahmed, Z.; Shafiq, H. Therapeutic potential of flavonoids and their mechanism of action against microbial and viral infections—A review. Food Res. Int. 2015, 77, 221–235. [Google Scholar] [CrossRef]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chem.-Biol. Interact. 2020, 328, 109211. [Google Scholar] [CrossRef]
- Bardelčíková, A.; Miroššay, A.; Šoltýs, J.; Mojžiš, J. Therapeutic and prophylactic effect of flavonoids in post-COVID-19 therapy. Phytother. Res. 2022, 36, 2042–2060. [Google Scholar] [CrossRef] [PubMed]
- Zakaryan, H.; Arabyan, E.; Oo, A.; Zandi, K. Flavonoids: Promising natural compounds against viral infections. Arch. Virol. 2017, 162, 2539–2551. [Google Scholar] [CrossRef]
- Alzaabi, M.M.; Hamdy, R.; Ashmawy, N.S.; Hamoda, A.M.; Alkhayat, F.; Khademi, N.N.; Joud, A.; Abo, S.M.; El-Keblawy, A.A.; Soliman, S.S. Flavonoids are promising safe therapy against COVID-19. Phytochem. Rev. 2022, 21, 291–312. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Farid, M.M.; Mostafa, A.; Ragheb, A.Y.; Mahmoud, S.H.; Shehata, M.; Shama, N.M.A.; GabAllah, M.; Mostafa-Hedeab, G.; Marzouk, M.M. Naturally available flavonoid aglycones as potential antiviral drug candidates against SARS-CoV-2. Molecules 2021, 26, 6559. [Google Scholar] [CrossRef]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Li, S.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzym. Inhib. Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Du, A.; Zheng, R.; Disoma, C.; Li, S.; Chen, Z.; Li, S.; Liu, P.; Zhou, Y.; Shen, Y.; Liu, S. Epigallocatechin-3-gallate, an active ingredient of Traditional Chinese Medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int. J. Biol. Macromol. 2021, 176, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zhu, G.-H.; Zhang, Y.-N.; Hu, Q.; Wang, H.-N.; Yu, H.-N.; Qin, X.-Y.; Guan, X.-Q.; Xiang, Y.-W.; Tang, H. Flavonoids in Ampelopsis grossedentata as covalent inhibitors of SARS-CoV-2 3CLpro: Inhibition potentials, covalent binding sites and inhibitory mechanisms. Int. J. Biol. Macromol. 2021, 187, 976–987. [Google Scholar] [CrossRef]
- Rizzuti, B.; Grande, F.; Conforti, F.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Ortega-Alarcon, D.; Vega, S.; Reyburn, H.T.; Abian, O.; Velazquez-Campoy, A. Rutin is a low micromolar inhibitor of SARS-CoV-2 main protease 3CLpro: Implications for drug design of quercetin analogs. Biomedicines 2021, 9, 375. [Google Scholar] [CrossRef]
- Zandi, K.; Musall, K.; Oo, A.; Cao, D.; Liang, B.; Hassandarvish, P.; Lan, S.; Slack, R.L.; Kirby, K.A.; Bassit, L. Baicalein and baicalin inhibit SARS-CoV-2 RNA-dependent-RNA polymerase. Microorganisms 2021, 9, 893. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, Y.e.; Zhang, Y.; Zhang, R.; Zhu, C.; Fan, L.; Pei, G.; Zhang, B.; Shi, Y. Baicalein inhibits SARS-CoV-2/VSV replication with interfering mitochondrial oxidative phosphorylation in a mPTP dependent manner. Signal Transduct. Target. Ther. 2020, 5, 266. [Google Scholar] [CrossRef]
- Bongini, P.; Trezza, A.; Bianchini, M.; Spiga, O.; Niccolai, N. A possible strategy to fight COVID-19: Interfering with spike glycoprotein trimerization. Biochem. Biophys. Res. Commun. 2020, 528, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Raghuvanshi, R.; Ceylan, F.D.; Bolling, B.W. Quercetin and its metabolites inhibit recombinant human angiotensin-converting enzyme 2 (ACE2) activity. J. Agric. Food Chem. 2020, 68, 13982–13989. [Google Scholar] [CrossRef] [PubMed]
- Güler, H.I.; Şal, F.A.; Can, Z.; Kara, Y.; Yildiz, O.; Beldüz, A.O.; Canakci, S.; Kolayli, S. Targeting CoV-2 spike RBD and ACE-2 interaction with flavonoids of Anatolian propolis by in silico and in vitro studies in terms of possible COVID-19 therapeutics. Turk. J. Biol. 2021, 45, 530–548. [Google Scholar] [CrossRef]
- Biagioli, M.; Marchianò, S.; Roselli, R.; Di Giorgio, C.; Bellini, R.; Bordoni, M.; Gidari, A.; Sabbatini, S.; Francisci, D.; Fiorillo, B. Discovery of a AHR pelargonidin agonist that counter-regulates Ace2 expression and attenuates ACE2-SARS-CoV-2 interaction. Biochem. Pharmacol. 2021, 188, 114564. [Google Scholar] [CrossRef]
- Zhan, Y.; Ta, W.; Tang, W.; Hua, R.; Wang, J.; Wang, C.; Lu, W. Potential antiviral activity of isorhamnetin against SARS-CoV-2 spike pseudotyped virus in vitro. Drug Dev. Res. 2021, 82, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Ding, Y.; Wang, Y.; Liang, P.; Zhang, L.; Liu, R. Oroxylin A is a severe acute respiratory syndrome coronavirus 2-spiked pseudotyped virus blocker obtained from Radix Scutellariae using angiotensin-converting enzyme II/cell membrane chromatography. Phytother. Res. 2021, 35, 3194–3204. [Google Scholar] [CrossRef]
- Henss, L.; Auste, A.; Schürmann, C.; Schmidt, C.; von Rhein, C.; Mühlebach, M.D.; Schnierle, B.S. The green tea catechin epigallocatechin gallate inhibits SARS-CoV-2 infection. J. Gen. Virol. 2021, 102, 001574. [Google Scholar] [CrossRef]
- Liu, J.; Meng, J.; Li, R.; Jiang, H.; Fu, L.; Xu, T.; Zhu, G.-Y.; Zhang, W.; Jiang, Z.-H.; Yang, Z.-F. Integrated Network Pharmacology Analysis, Molecular Docking, LC-MS Analysis and Bioassays Revealed the Potential Active Ingredients and Underlying Mechanism of Scutellariae Radix for COVID-19. Front. Plant Sci. 2022, 13, 988655. [Google Scholar] [CrossRef]
- Guo, Y.; Meng, J.-R.; Liu, J.-Z.; Xu, T.; Zheng, Z.-Y.; Jiang, Z.-H.; Bai, L.-P. Synthesis and Biological Evaluation of Honokiol Derivatives Bearing 3-((5-phenyl-1, 3, 4-oxadiazol-2-yl) methyl) oxazol-2 (3H)-ones as Potential Viral Entry Inhibitors against SARS-CoV-2. Pharmaceuticals 2021, 14, 885. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Meng, J.; Tian, W.; Liu, J.; Hu, X.; Jiang, Z.-H.; Zhang, W.; Li, Y.; Bai, L.-P. I2-Catalyzed Carbonylation of α-Methylene Ketones to Synthesize 1, 2-Diaryl Diketones and Antiviral Quinoxalines in One Pot. ACS Omega 2021, 7, 1380–1394. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Meng, J.-R.; Cheng, W.; Liu, J.-Z.; Chu, J.; Zhang, Q.; Ma, N.; Bai, L.-P.; Guo, Y. Discovery of honokiol thioethers containing 1, 3, 4-oxadiazole moieties as potential α-glucosidase and SARS-CoV-2 entry inhibitors. Bioorg. Med. Chem. 2022, 67, 116838. [Google Scholar] [CrossRef]
- Fährrolfes, R.; Bietz, S.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Otto, T.; Volkamer, A.; Rarey, M. Proteins Plus: A web portal for structure analysis of macromolecules. Nucleic Acids Res. 2017, 45, W337–W343. [Google Scholar] [CrossRef] [Green Version]
- Schöning-Stierand, K.; Diedrich, K.; Fährrolfes, R.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Steinegger, R.; Rarey, M. Proteins Plus: Interactive analysis of protein–ligand binding interfaces. Nucleic Acids Res. 2020, 48, W48–W53. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, H.; Watashi, K.; Saso, W.; Shionoya, K.; Iwanami, S.; Hirokawa, T.; Shirai, T.; Kanaya, S.; Ito, Y.; Kim, K.S. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. Iscience 2021, 24, 102367. [Google Scholar] [CrossRef]
- Chen, C.Z.; Xu, M.; Pradhan, M.; Gorshkov, K.; Petersen, J.D.; Straus, M.R.; Zhu, W.; Shinn, P.; Guo, H.; Shen, M. Identifying SARS-CoV-2 entry inhibitors through drug repurposing screens of SARS-S and MERS-S pseudotyped particles. ACS Pharmacol. Transl. Sci. 2020, 3, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Indrayanto, G.; Putra, G.S.; Suhud, F. Validation of in-vitro bioassay methods: Application in herbal drug research. Profiles Drug Subst. Excip. Relat. Methodol. 2021, 46, 273–307. [Google Scholar] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Solopov, P.; Biancatelli, R.C.; Dimitropoulou, C. Dietary phytoestrogens ameliorate hydrochloric acid-induced chronic lung injury and pulmonary fibrosis in mice. Nutrients 2021, 13, 3599. [Google Scholar] [CrossRef] [PubMed]






| No. | Flavonoids | CC0 a (μM) | TC50 b (μM) | IC50 c (μM) | SI d |
|---|---|---|---|---|---|
| 1 | Chrysin | 100 | 168.50 ± 12.06 | >100 | <1.69 |
| 2 | Apigenin | 50 | 90.88 ± 3.13 | >50 | <1.82 |
| 3 | Luteolin | 200 | >300 | 17.45 ± 4.46 | >17.19 |
| 4 | Vitexin | 300 | >500 | >300 | ND |
| 5 | Orientin | 300 | >500 | 63.73 ± 9.61 | >7.85 |
| 6 | Vitexin-2″-O-rhamnoside | 300 | >500 | >300 | ND |
| 7 | Baicalein | 200 | >500 | 51.04 ± 6.48 | >9.80 |
| 8 | Oroxylin A | 100 | >300 | 37.12 ± 2.58 | >8.08 |
| 9 | Scutellarein | 200 | >300 | 29.26 ± 0.74 | >10.25 |
| 10 | Oroxin A | 300 | >300 | 84.61 ± 10.81 | >3.55 |
| 11 | Oroxin B | 300 | >300 | 128.70 ± 19.03 | >2.33 |
| 12 | Baicalin | 300 | >500 | 109.63 ± 11.02 | >4.56 |
| 13 | Wogonoside | 300 | >500 | 119.07 ± 21.00 | >4.20 |
| 14 | Kaempferol | 200 | >200 | 34.65 ± 7.69 | >5.77 |
| 15 | Quercetin | 300 | >500 | 17.00 ± 3.42 | >29.41 |
| 16 | Myricetin | 300 | >500 | 10.27 ± 2.32 | >48.69 |
| 17 | Astragalin | 300 | >500 | 133.05 ± 16.19 | >3.76 |
| 18 | Isoquercitrin | 300 | >500 | 94.39 ± 1.59 | >5.30 |
| 19 | Hyperoside | 300 | >500 | 83.12 ± 10.65 | >6.02 |
| 20 | Rutin | 300 | >500 | 146.87 ± 21.11 | >3.40 |
| 21 | (±)-Catechin | 300 | >500 | 156.80 ± 12.00 | >3.19 |
| 22 | (-)-Epicatechin (EC) | 300 | >500 | 145.77 ± 20.82 | >3.43 |
| 23 | (-)-Epigallocatechin (EGC) | 200 | 453.10 ± 4.55 | 74.21 ± 10.72 | 6.11 |
| 24 | (-)-Epicatechin gallate (ECG) | 300 | >500 | 58.77 ± 3.25 | >8.51 |
| 25 | (-)-Epigallocatechin gallate (EGCG) | 300 | >500 | 31.26 ± 7.40 | >15.99 |
| 26 | (-)-Gallocatechin gallate (GCG) | 300 | >500 | 43.41 ± 2.93 | >11.52 |
| 27 | Procyanidin B-1 | 300 | >500 | >300 | ND |
| 28 | Procyanidin B-2 | 300 | >500 | >300 | ND |
| 29 | Procyanidin B-3 | 300 | >500 | >300 | ND |
| 30 | Procyanidin B-4 | 300 | >500 | 151.37 ± 28.08 | >3.30 |
| 31 | Procyanidin C-1 | 300 | >500 | 172.63 ± 20.74 | >2.90 |
| Positive control | Cepharanthine | 6.25 | 11.54 ± 1.39 | 1.30 ± 0.18 | 8.88 |
| Number | Volume Å3 | Drug Score | Site Center (X, Y, Z) |
|---|---|---|---|
| Pocket 1 | 216.45 | 0.59 | −22.987082, 21.412573, 35.788383 |
| Pocket 2 | 183.42 | 0.46 | −24.487082, 22.662573, 19.788383 |
| Pocket 3 | 181.95 | 0.45 | −38.987082, 42.412573, 12.288383 |
| Pocket 4 | 148.80 | 0.31 | −24.987082, 17.662573, 31.288383 |
| Pocket 5 | 109.63 | 0.16 | −35.637121, 22.231293, 7.207497 |
| No. | Flavonoids | IC50 Values of Human Normal Cell Lines (μM) | ||
|---|---|---|---|---|
| Beas-2B | LO2 | HEK 293 | ||
| 3 | Luteolin | >300 | 182.68 ± 10.26 | >300 |
| 5 | Orientin | >500 | >500 | >500 |
| 7 | Baicalein | 493.75 ± 9.64 | >500 | >500 |
| 8 | Oroxylin A | >300 | >300 | >300 |
| 9 | Scutellarein | >300 | >300 | >300 |
| 10 | Oroxin A | >300 | >300 | 257.80 ± 10.57 |
| 11 | Oroxin B | >300 | >300 | >300 |
| 12 | Baicalin | >500 | >500 | >500 |
| 13 | Wogonoside | >500 | >500 | >500 |
| 14 | Kaempferol | >200 | >200 | >200 |
| 15 | Quercetin | >500 | >500 | >500 |
| 16 | Myricetin | >500 | >500 | >500 |
| 17 | Astragalin | >500 | >500 | >500 |
| 18 | Isoquercitrin | >500 | >500 | >500 |
| 19 | Hyperoside | >500 | >500 | >500 |
| 20 | Rutin | >500 | >500 | >500 |
| 21 | (±)-Catechin | >500 | >500 | >500 |
| 22 | (-)-Epicatechin (EC) | >500 | >500 | >500 |
| 23 | (-)-Epigallocatechin (EGC) | 369.00 ± 1.70 | 483.33 ± 1.52 | >500 |
| 24 | (-)-Epicatechin gallate (ECG) | >500 | >500 | >500 |
| 25 | (-)-Epigallocatechin gallate (EGCG) | >500 | 430.40 ± 6.98 | >500 |
| 26 | (-)-Gallocatechin gallate (GCG) | >500 | 432.93 ± 8.96 | >500 |
| 30 | Procyanidin B-4 | >500 | >500 | >500 |
| 31 | Procyanidin C-1 | >500 | >500 | >500 |
| Positive control | Cepharanthine | 26.13 ± 1.08 | 16.06 ± 0.13 | 24.30 ± 0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, J.-R.; Liu, J.; Fu, L.; Shu, T.; Yang, L.; Zhang, X.; Jiang, Z.-H.; Bai, L.-P. Anti-Entry Activity of Natural Flavonoids against SARS-CoV-2 by Targeting Spike RBD. Viruses 2023, 15, 160. https://doi.org/10.3390/v15010160
Meng J-R, Liu J, Fu L, Shu T, Yang L, Zhang X, Jiang Z-H, Bai L-P. Anti-Entry Activity of Natural Flavonoids against SARS-CoV-2 by Targeting Spike RBD. Viruses. 2023; 15(1):160. https://doi.org/10.3390/v15010160
Chicago/Turabian StyleMeng, Jie-Ru, Jiazheng Liu, Lu Fu, Tong Shu, Lingzhi Yang, Xueji Zhang, Zhi-Hong Jiang, and Li-Ping Bai. 2023. "Anti-Entry Activity of Natural Flavonoids against SARS-CoV-2 by Targeting Spike RBD" Viruses 15, no. 1: 160. https://doi.org/10.3390/v15010160
APA StyleMeng, J.-R., Liu, J., Fu, L., Shu, T., Yang, L., Zhang, X., Jiang, Z.-H., & Bai, L.-P. (2023). Anti-Entry Activity of Natural Flavonoids against SARS-CoV-2 by Targeting Spike RBD. Viruses, 15(1), 160. https://doi.org/10.3390/v15010160