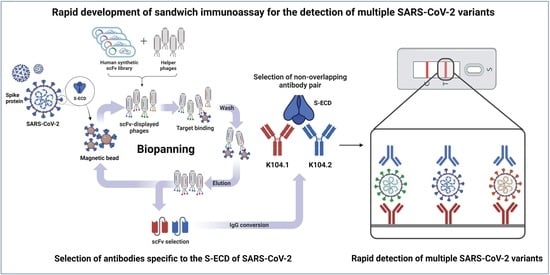
Development and Characterization of Phage Display-Derived Monoclonal Antibodies to the S2 Domain of Spike Proteins of Wild-Type SARS-CoV-2 and Multiple Variants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of scFvs Using Phage Display Technology
2.2. Phage ELISA
2.3. DNA Cloning
2.4. Cell Culture
2.5. Preparation of SARS-CoV-2 Spike Protein-Specific IgG-Based mAbs
2.6. Surface Plasmon Resonance (SPR)
2.7. ELISA
2.8. Competition ELISA
2.9. Sandwich Immunoassay
2.10. Statistical Validation of the Sandwich Immunoassay
3. Results
3.1. Selection and Biochemical Characterization of SARS-CoV-2 Spike Protein-Specific IgG-Based mAbs
3.2. Selection of an Antibody Pair Comprising of Antibodies with Different Epitopes and High Binding Affinity to SARS-CoV-2 Spike Protein
3.3. Development and Characterization of Sandwich Immunoassay for the Detection of Wild-Type SARS-CoV-2 Spike Protein
3.4. Detection of the Spike Proteins of Multiple SARS-CoV-2 Variants and Other Coronaviruses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chatterjee, P.; Nagi, N.; Agarwal, A.; Das, B.; Banerjee, S.; Sarkar, S.; Gupta, N.; Gangakhedkar, R.R. The 2019 novel coronavirus disease (COVID-19) pandemic: A review of the current evidence. Indian J. Med. Res. 2020, 151, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, S.; Wu, W.; Geng, S.; Mao, M. Distinct mutations and lineages of SARS-CoV-2 virus in the early phase of COVID-19 pandemic and subsequent 1-year global expansion. J. Med. Virol. 2022, 94, 2035–2049. [Google Scholar] [CrossRef] [PubMed]
- Khateeb, J.; Li, Y.; Zhang, H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care 2021, 25, 244. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). COVID-19 Weekly Epidemiological Update. 2022. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---4-january-2023 (accessed on 5 January 2023).
- Guan, W.J.; Chen, R.C.; Zhong, N.S. Strategies for the prevention and management of coronavirus disease 2019. Eur. Respir. J. 2020, 55, 2000597. [Google Scholar] [CrossRef] [Green Version]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Jarrom, D.; Elston, L.; Washington, J.; Prettyjohns, M.; Cann, K.; Myles, S.; Groves, P. Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: A rapid systematic review. BMJ Evid. Based Med. 2022, 27, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Meng, H.; Liu, H.; Ye, Q. Advances in laboratory detection methods and technology application of SARS-CoV-2. J. Med. Virol. 2022, 94, 1357–1365. [Google Scholar] [CrossRef]
- Dos Santos, P.G.; Vieira, H.; Wietholter, V.; Gallina, J.P.; Andrade, T.R.; Marinowic, D.R.; Zanirati, G.G.; da Costa, J.C. When to test for COVID-19 using real-time reverse transcriptase polymerase chain reaction: A systematic review. Int. J. Infect. Dis. 2022, 123, 58–69. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Mina, M.J.; Parker, R.; Larremore, D.B. Rethinking Covid-19 test sensitivity—A strategy for containment. N. Engl. J. Med. 2020, 383, e120. [Google Scholar] [CrossRef]
- Yokota, I.; Shane, P.Y.; Okada, K.; Unoki, Y.; Yang, Y.; Iwasaki, S.; Fujisawa, S.; Nishida, M.; Teshima, T. A novel strategy for SARS-CoV-2 mass screening with quantitative antigen testing of saliva: A diagnostic accuracy study. Lancet Microbe 2021, 2, e397–e404. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, K.A.; Dewald, F.; Heger, E.; Gieselmann, L.; Vanshylla, K.; Wirtz, M.; Kleipass, F.; Johannis, W.; Schommers, P.; Gruell, H.; et al. Evaluation of a new spike (S)-protein-based commercial immunoassay for the detection of anti-SARS-CoV-2 IgG. Microorganisms 2021, 9, 733. [Google Scholar] [CrossRef] [PubMed]
- Favresse, J.; Cadrobbi, J.; Eucher, C.; Elsen, M.; Laffineur, K.; Dogné, J.-M.; Douxfils, J. Clinical performance of three fully automated anti-SARS-CoV-2 immunoassays targeting the nucleocapsid or spike proteins. J. Med. Virol. 2021, 93, 2262–2269. [Google Scholar] [CrossRef] [PubMed]
- Maia, R.; Carvalho, V.; Faria, B.; Miranda, I.; Catarino, S.; Teixeira, S.; Lima, R.; Minas, G.; Ribeiro, J. Diagnosis methods for COVID-19: A systematic review. Micromachines 2022, 13, 1349. [Google Scholar] [CrossRef] [PubMed]
- Pinals, R.L.; Ledesma, F.; Yang, D.; Navarro, N.; Jeong, S.; Pak, J.E.; Kuo, L.; Chuang, Y.-C.; Cheng, Y.-W.; Sun, H.-Y.; et al. Rapid SARS-CoV-2 spike protein detection by carbon nanotube-based near-infrared nanosensors. Nano Lett. 2021, 21, 2272–2280. [Google Scholar] [CrossRef] [PubMed]
- Ahmadivand, A.; Gerislioglu, B.; Ramezani, Z.; Kaushik, A.; Manickam, P.; Ghoreishi, S.A. Functionalized terahertz plasmonic metasensors: Femtomolar-level detection of SARS-CoV-2 spike proteins. Biosens. Bioelectron. 2021, 177, 112971. [Google Scholar] [CrossRef]
- Idili, A.; Parolo, C.; Alvarez-Diduk, R.; Merkoçi, A. Rapid and efficient detection of the SARS-CoV-2 spike protein using an electrochemical aptamer-based sensor. ACS Sens. 2021, 6, 3093–3101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Juhas, M.; Kwok, C.K. Aptamers targeting SARS-COV-2: A promising tool to fight against COVID-19. Trends Biotechnol. 2022. [Google Scholar] [CrossRef]
- Leuzinger, K.; Roloff, T.; Egli, A.; Hirsch, H.H. Impact of SARS-CoV-2 Omicron on rapid antigen testing developed for early-pandemic SARS-CoV-2 variants. Microbiol. Spectr. 2022, 10, e0200622. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-F.; Xu, W.; Liu, S.-W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Pizzato, M.; Baraldi, C.; Boscato Sopetto, G.; Finozzi, D.; Gentile, C.; Gentile, M.D.; Marconi, R.; Paladino, D.; Raoss, A.; Riedmiller, I.; et al. SARS-CoV-2 and the host cell: A tale of interactions. Front. Virol. 2022, 1, 815388. [Google Scholar] [CrossRef]
- Triveri, A.; Serapian, S.A.; Marchetti, F.; Doria, F.; Pavoni, S.; Cinquini, F.; Moroni, E.; Rasola, A.; Frigerio, F.; Colombo, G. SARS-CoV-2 Spike Protein Mutations and Escape from Antibodies: A Computational Model of Epitope Loss in Variants of Concern. J. Chem. Inf. Model. 2021, 61, 4687–4700. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Canziani, G.A.; Carter, E.P.; Chaiken, I. The case for S2: The potential benefits of the S2 subunit of the SARS-CoV-2 spike protein as an immunogen in fighting the COVID-19 pandemic. Front. Immunol. 2021, 12, 637651. [Google Scholar] [CrossRef]
- Yang, H.Y.; Kang, K.J.; Chung, J.E.; Shim, H. Construction of a large synthetic human scFv library with six diversified CDRs and high functional diversity. Mol. Cells 2009, 27, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, J.W.; Yang, H.R.; Song, S.-W.; Lee, S.-J.; Jeon, Y.; Ju, A.; Lee, N.; Kim, M.-G.; Kim, M.; et al. A fully-human antibody specifically targeting a membrane-bound fragment of CADM1 potentiates the T cell-mediated death of human small-cell lung cancer cells. Int. J. Mol. Sci. 2022, 23, 6895. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.B.; Kim, J.W.; Heo, K.; Kim, H.J.; Yun, S.; Lee, H.S.; Shin, H.G.; Shim, H.; Yu, H.; Kim, Y.-H.; et al. An internalizing antibody targeting of cell surface GRP94 effectively suppresses tumor angiogenesis of colorectal cancer. Biomed. Pharmacother. 2022, 150, 113051. [Google Scholar] [CrossRef]
- Shah, K.; Maghsoudlou, P. Enzyme-linked immunosorbent assay (ELISA): The basics. Br. J. Hosp. Med. 2016, 77, C98–C101. [Google Scholar] [CrossRef]
- Xiao, M.; Tian, F.; Liu, X.; Zhou, Q.; Pan, J.; Luo, Z.; Yang, M.; Yi, C. Virus detection: From state-of-the-art laboratories to smartphone-based point-of-care testing. Adv. Sci. 2022, 9, 2105904. [Google Scholar] [CrossRef]
- Zai, J.; Yi, K.; Xie, L.; Zhu, J.; Feng, X.; Li, Y. Dual monoclonal antibody-based sandwich ELISA for detection of in vitro packaged Ebola virus. Diagn. Pathol. 2018, 13, 96. [Google Scholar] [CrossRef]
- Kuang, H.; Wang, W.; Xu, L.; Ma, W.; Liu, L.; Wang, L.; Xu, C. Monoclonal antibody-based sandwich ELISA for the detection of staphylococcal enterotoxin A. Int. J. Environ. Res. Public Health 2013, 10, 1598–1608. [Google Scholar] [CrossRef]
- Kadian, N.; Raju, K.S.R.; Rashid, M.; Malik, M.Y.; Taneja, I.; Wahajuddin, M. Comparative assessment of bioanalytical method validation guidelines for pharmaceutical industry. J. Pharm. Biomed. Anal. 2016, 126, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19). In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK554776/ (accessed on 3 November 2022).
- Keni, R.; Alexander, A.; Nayak, P.G.; Mudgal, J.; Nandakumar, K. COVID-19: Emergence, spread, possible treatments, and global Burden. Front. Public Health 2020, 8, 216. [Google Scholar] [CrossRef] [PubMed]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The disease and tools for detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [Green Version]
- Frenzel, A.; Schirrmann, T.; Hust, M. Phage display-derived human antibodies in clinical development and therapy. MAbs 2016, 8, 1177–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, T.S.; Hust, M.; Visai, L. Editorial: Recent advances in recombinant antibody therapeutics and diagnostics for infectious diseases. Front. Public Health 2022, 10, 876889. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A. Monoclonal antibodies in diagnosis and therapy. Science 1991, 252, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, A.; Chen, Y.; Sun, Y.; Du, Y.; Wang, X.; Ding, P.; Jia, R.; Wang, Y.; Zhang, G. Development of a colloidal gold-based immunochromatographic strip for rapid detection of severe acute respiratory syndrome coronavirus 2 spike protein. Front. Immunol. 2021, 12, 635677. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A.; et al. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020, 6, 2046–2052. [Google Scholar] [CrossRef]
- Maeda, R.; Fujita, J.; Konishi, Y.; Kazuma, Y.; Yamazaki, H.; Anzai, I.; Watanabe, T.; Yamaguchi, K.; Kasai, K.; Nagata, K.; et al. A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron. Commun. Biol. 2022, 5, 669. [Google Scholar] [CrossRef]
- Svobodova, M.; Skouridou, V.; Jauset-Rubio, M.; Viéitez, I.; Fernández-Villar, A.; Cabrera Alvargonzalez, J.J.; Poveda, E.; Bofill, C.B.; Sans, T.; Bashammakh, A.; et al. Aptamer sandwich assay for the detection of SARS-CoV-2 spike protein antigen. ACS Omega 2021, 6, 35657–35666. [Google Scholar] [CrossRef]
- Di Domenico, M.; De Rosa, A.; Boccellino, M. Detection of SARS-COV-2 proteins using an ELISA test. Diagnostics 2021, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Valtanen, P.; Hope, C.M.; Masavuli, M.G.; Yeow, A.E.L.; Balachandran, H.; Mekonnen, Z.A.; AlDelfi, Z.; Abayasingam, A.; Agapiou, D.; Stella, A.O.; et al. SARS-CoV-2 Omicron variant escapes neutralizing antibodies and T cell responses more efficiently than other variants in mild COVID-19 convalescents. Cell Rep. Med. 2022, 3, 100651. [Google Scholar] [CrossRef] [PubMed]
- Kudriavtsev, A.V.; Vakhrusheva, A.V.; Novoseletsky, V.N.; Bozdaganyan, M.E.; Shaitan, K.V.; Kirpichnikov, M.P.; Sokolova, O.S. Immune escape associated with RBD Omicron mutations and SARS-CoV-2 evolution dynamics. Viruses 2022, 14, 1603. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, W. Fast-spreading SARS-CoV-2 variants: Challenges to and new design strategies of COVID-19 vaccines. Signal Transduct. Target. Ther. 2021, 6, 226. [Google Scholar] [CrossRef]
- Farinholt, T.; Doddapaneni, H.; Qin, X.; Menon, V.; Meng, Q.; Metcalf, G.; Chao, H.; Gingras, M.-C.; Avadhanula, V.; Farinholt, P.; et al. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med. 2021, 19, 255. [Google Scholar] [CrossRef]





| mAb | 1KD (nM) | 2Ka (1/M−1s−1) | 3Kd (s−1) |
|---|---|---|---|
| K104.1 | 1.28 | 3.18 105 | 4.08 10−4 |
| K104.2 | 1.90 | 4.04 105 | 7.68 10−4 |
| K104.3 | 8.22 | 2.87 105 | 2.36 10−3 |
| K104.4 | 12.21 | 3.15 104 | 3.85 10−4 |
| Spiked Level (ng/mL) | Intra-Assay (n = 6) | Inter-Assay (n = 6) | ||||
|---|---|---|---|---|---|---|
| Mean ± SD (ng/mL) | Recovery (%) | CV (%) | Mean ± SD (ng/mL) | Recovery (%) | CV (%) | |
| 50 | 51.12 3.58 | 102.25 | 7.17 | 48.97 3.57 | 97.95 | 7.28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.W.; Cho, A.H.; Shin, H.G.; Jang, S.H.; Cho, S.Y.; Lee, Y.R.; Lee, S. Development and Characterization of Phage Display-Derived Monoclonal Antibodies to the S2 Domain of Spike Proteins of Wild-Type SARS-CoV-2 and Multiple Variants. Viruses 2023, 15, 174. https://doi.org/10.3390/v15010174
Kim JW, Cho AH, Shin HG, Jang SH, Cho SY, Lee YR, Lee S. Development and Characterization of Phage Display-Derived Monoclonal Antibodies to the S2 Domain of Spike Proteins of Wild-Type SARS-CoV-2 and Multiple Variants. Viruses. 2023; 15(1):174. https://doi.org/10.3390/v15010174
Chicago/Turabian StyleKim, Ji Woong, Ah Hyun Cho, Ha Gyeong Shin, Sung Hoon Jang, Su Yeon Cho, Ye Rim Lee, and Sukmook Lee. 2023. "Development and Characterization of Phage Display-Derived Monoclonal Antibodies to the S2 Domain of Spike Proteins of Wild-Type SARS-CoV-2 and Multiple Variants" Viruses 15, no. 1: 174. https://doi.org/10.3390/v15010174
APA StyleKim, J. W., Cho, A. H., Shin, H. G., Jang, S. H., Cho, S. Y., Lee, Y. R., & Lee, S. (2023). Development and Characterization of Phage Display-Derived Monoclonal Antibodies to the S2 Domain of Spike Proteins of Wild-Type SARS-CoV-2 and Multiple Variants. Viruses, 15(1), 174. https://doi.org/10.3390/v15010174