Risk Factors of Severe COVID-19: A Review of Host, Viral and Environmental Factors
Abstract
:1. Introduction
2. Host Factors
2.1. Age
2.2. Human Genetic Variation
2.3. Sex and Pregnancy
2.4. Comorbidities
2.4.1. Non-Communicable Diseases (NCDs)
2.4.2. Coinfections/Superinfections
2.5. Frailty
2.6. Microbiota
2.7. Immunological History
2.7.1. Previous SARS-CoV-2 Infection
2.7.2. Cross-Reactive Immunity from Other Infections
2.8. Lifestyle
2.8.1. Physical Activity
2.8.2. Alcohol Consumption
2.8.3. Smoking
2.8.4. Diet and Nutrition
3. Viral Factors
3.1. Viral Genetic Variation
3.2. Infecting Dose (Inoculum Size)
4. Environmental Factors
4.1. Socioeconomic Factors
4.2. Air Pollution
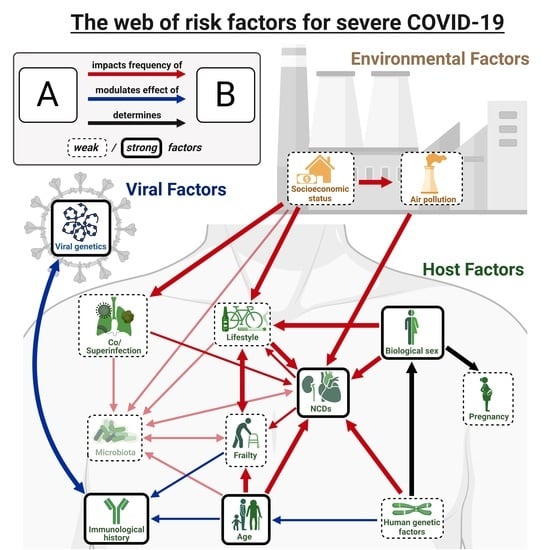
5. Interactions between Effects
5.1. Interactions between Risk Factors of Severe COVID-19
5.2. Direct and Indirect Effects of COVID-19 on the Risk Factors of Severe Disease
6. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Relative Risk Measures Used to Identify COVID-19 Risk Factors
References
- Vegivinti, C.T.R.; Evanson, K.W.; Lyons, H.; Akosman, I.; Barrett, A.; Hardy, N.; Kane, B.; Keesari, P.R.; Pulakurthi, Y.S.; Sheffels, E.; et al. Efficacy of Antiviral Therapies for COVID-19: A Systematic Review of Randomized Controlled Trials. BMC Infect. Dis. 2022, 22, 107. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. National Institutes of Health COVID-19 Treatment Guidelines Panel. Available online: https://www.covid19treatmentguidelines.nih.gov/ (accessed on 9 December 2022).
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated with Acute Respiratory Distress Syndrome and Death in Patients with Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; Yu, T.; et al. Clinical Course and Outcomes of Critically Ill Patients with SARS-CoV-2 Pneumonia in Wuhan, China: A Single-Centered, Retrospective, Observational Study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pijls, B.G.; Jolani, S.; Atherley, A.; Derckx, R.T.; Dijkstra, J.I.R.; Franssen, G.H.L.; Hendriks, S.; Richters, A.; Venemans-Jellema, A.; Zalpuri, S.; et al. Demographic Risk Factors for COVID-19 Infection, Severity, ICU Admission and Death: A Meta-Analysis of 59 Studies. BMJ Open 2021, 11, e044640. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Li, S.; Yu, H.; Wang, P.; Zhang, Y.; Chen, Z.; Li, Y.; Cheng, L.; Li, W.; Jia, H.; et al. Epidemiological, Comorbidity Factors with Severity and Prognosis of COVID-19: A Systematic Review and Meta-Analysis. Aging 2020, 12, 12493–12503. [Google Scholar] [CrossRef] [PubMed]
- Verity, R.; Okell, L.C.; Dorigatti, I.; Winskill, P.; Whittaker, C.; Imai, N.; Cuomo-Dannenburg, G.; Thompson, H.; Walker, P.G.T.; Fu, H.; et al. Estimates of the Severity of Coronavirus Disease 2019: A Model-Based Analysis. Lancet Infect. Dis. 2020, 20, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Salje, H.; Tran Kiem, C.; Lefrancq, N.; Courtejoie, N.; Bosetti, P.; Paireau, J.; Andronico, A.; Hozé, N.; Richet, J.; Dubost, C.-L.; et al. Estimating the Burden of SARS-CoV-2 in France. Science 2020, 369, 208–211. [Google Scholar] [CrossRef]
- O’Driscoll, M.; Ribeiro Dos Santos, G.; Wang, L.; Cummings, D.A.T.; Azman, A.S.; Paireau, J.; Fontanet, A.; Cauchemez, S.; Salje, H. Age-Specific Mortality and Immunity Patterns of SARS-CoV-2. Nature 2021, 590, 140–145. [Google Scholar] [CrossRef]
- Du, P.; Li, D.; Wang, A.; Shen, S.; Ma, Z.; Li, X. A Systematic Review and Meta-Analysis of Risk Factors Associated with Severity and Death in COVID-19 Patients. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, e6660930. [Google Scholar] [CrossRef]
- Booth, A.; Reed, A.B.; Ponzo, S.; Yassaee, A.; Aral, M.; Plans, D.; Labrique, A.; Mohan, D. Population Risk Factors for Severe Disease and Mortality in COVID-19: A Global Systematic Review and Meta-Analysis. PLoS ONE 2021, 16, e0247461. [Google Scholar] [CrossRef]
- Ebinger, J.E.; Achamallah, N.; Ji, H.; Claggett, B.L.; Sun, N.; Botting, P.; Nguyen, T.-T.; Luong, E.; Kim, E.H.; Park, E.; et al. Pre-Existing Traits Associated with COVID-19 Illness Severity. PLoS ONE 2020, 15, e0236240. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Vega, M.F.; Salinas-Escudero, G.; García-Peña, C.; Gutiérrez-Robledo, L.M.; Parra-Rodríguez, L. Early Estimation of the Risk Factors for Hospitalization and Mortality by COVID-19 in Mexico. PLoS ONE 2020, 15, e0238905. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.L.; Rowe, J.H.; Garcia-de-Alba, C.; Kim, C.F.; Sharpe, A.H.; Haigis, M.C. The Aging Lung: Physiology, Disease, and Immunity. Cell 2021, 184, 1990–2019. [Google Scholar] [CrossRef]
- Sanchez-Vazquez, R.; Guío-Carrión, A.; Zapatero-Gaviria, A.; Martínez, P.; Blasco, M.A. Shorter Telomere Lengths in Patients with Severe COVID-19 Disease. Aging 2021, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chow, R.D.; Majety, M.; Chen, S. The Aging Transcriptome and Cellular Landscape of the Human Lung in Relation to SARS-CoV-2. Nat. Commun. 2021, 12, 4. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, Q.; Xia, X.; Liu, K.; Yu, Z.; Tao, W.; Gong, W.; Han, J.-D.J. Individual Variation of the SARS-CoV-2 Receptor ACE2 Gene Expression and Regulation. Aging Cell 2020, 19, e13168. [Google Scholar] [CrossRef]
- Schouten, L.R.; van Kaam, A.H.; Kohse, F.; Veltkamp, F.; Bos, L.D.; de Beer, F.M.; van Hooijdonk, R.T.; Horn, J.; Straat, M.; Witteveen, E.; et al. Age-Dependent Differences in Pulmonary Host Responses in ARDS: A Prospective Observational Cohort Study. Ann. Intensive Care 2019, 9, 55. [Google Scholar] [CrossRef] [Green Version]
- Inde, Z.; Croker, B.A.; Yapp, C.; Joshi, G.N.; Spetz, J.; Fraser, C.; Qin, X.; Xu, L.; Deskin, B.; Ghelfi, E.; et al. Age-Dependent Regulation of SARS-CoV-2 Cell Entry Genes and Cell Death Programs Correlates with COVID-19 Severity. Sci. Adv. 2021, 7, eabf8609. [Google Scholar] [CrossRef]
- Gheware, A.; Ray, A.; Rana, D.; Bajpai, P.; Nambirajan, A.; Arulselvi, S.; Mathur, P.; Trikha, A.; Arava, S.; Das, P.; et al. ACE2 Protein Expression in Lung Tissues of Severe COVID-19 Infection. Sci. Rep. 2022, 12, 4058. [Google Scholar] [CrossRef]
- Zheng, M. ACE2 and COVID-19 Susceptibility and Severity. Aging Dis. 2022, 13, 360–372. [Google Scholar] [CrossRef]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Schatzmann Peron, J.P.; Nakaya, H.I. ACE2 Expression Is Increased in the Lungs of Patients with Comorbidities Associated with Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.; Costa de Oliveira, S. The Impact of Angiotensin-Converting Enzyme 2 (ACE2) Expression Levels in Patients with Comorbidities on COVID-19 Severity: A Comprehensive Review. Microorganisms 2021, 9, 1692. [Google Scholar] [CrossRef] [PubMed]
- Chamsi-Pasha, M.A.R.; Shao, Z.; Tang, W.H.W. Angiotensin-Converting Enzyme 2 as a Therapeutic Target for Heart Failure. Curr. Heart Fail. Rep. 2014, 11, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Arendse, L.B.; Danser, A.H.J.; Poglitsch, M.; Touyz, R.M.; Burnett, J.C.; Llorens-Cortes, C.; Ehlers, M.R.; Sturrock, E.D. Novel Therapeutic Approaches Targeting the Renin-Angiotensin System and Associated Peptides in Hypertension and Heart Failure. Pharmacol. Rev. 2019, 71, 539–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Schuler, B.A.; Habermann, A.C.; Plosa, E.J.; Taylor, C.J.; Jetter, C.; Negretti, N.M.; Kapp, M.E.; Benjamin, J.T.; Gulleman, P.; Nichols, D.S.; et al. Age-Determined Expression of Priming Protease TMPRSS2 and Localization of SARS-CoV-2 in Lung Epithelium. J. Clin. Investig. 2021, 131, e140766. [Google Scholar] [CrossRef]
- Wang, A.; Chiou, J.; Poirion, O.B.; Buchanan, J.; Valdez, M.J.; Verheyden, J.M.; Hou, X.; Kudtarkar, P.; Narendra, S.; Newsome, J.M.; et al. Single-Cell Multiomic Profiling of Human Lungs Reveals Cell-Type-Specific and Age-Dynamic Control of SARS-CoV2 Host Genes. eLife 2020, 9, e62522. [Google Scholar] [CrossRef]
- Deng, X.; Hackbart, M.; Mettelman, R.C.; O’Brien, A.; Mielech, A.M.; Yi, G.; Kao, C.C.; Baker, S.C. Coronavirus Nonstructural Protein 15 Mediates Evasion of DsRNA Sensors and Limits Apoptosis in Macrophages. Proc. Natl. Acad. Sci. USA 2017, 114, E4251–E4260. [Google Scholar] [CrossRef] [Green Version]
- DeDiego, M.L.; Nieto-Torres, J.L.; Jiménez-Guardeño, J.M.; Regla-Nava, J.A.; Alvarez, E.; Oliveros, J.C.; Zhao, J.; Fett, C.; Perlman, S.; Enjuanes, L. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis. PLoS Pathog. 2011, 7, e1002315. [Google Scholar] [CrossRef]
- Danthi, P. Viruses and the Diversity of Cell Death. Annu. Rev. Virol. 2016, 3, 533–553. [Google Scholar] [CrossRef] [PubMed]
- Rea, I.M.; Alexander, H.D. Triple Jeopardy in Ageing: COVID-19, Co-Morbidities and Inflamm-Ageing. Ageing Res. Rev. 2022, 73, 101494. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Huuskonen, J.; Ojala, J.; Kauppinen, A.; Kaarniranta, K.; Suuronen, T. Activation of Innate Immunity System during Aging: NF-KB Signaling Is the Molecular Culprit of Inflamm-Aging. Ageing Res. Rev. 2008, 7, 83–105. [Google Scholar] [CrossRef] [PubMed]
- Brodin, P. Immune Determinants of COVID-19 Disease Presentation and Severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Rangel, F.A. Neutrophil-to-Lymphocyte Ratio and Lymphocyte-to-C-Reactive Protein Ratio in Patients with Severe Coronavirus Disease 2019 (COVID-19): A Meta-Analysis. J. Med. Virol. 2020, 92, 1733–1734. [Google Scholar] [CrossRef] [Green Version]
- Molony, R.D.; Nguyen, J.T.; Kong, Y.; Montgomery, R.R.; Shaw, A.C.; Iwasaki, A. Aging Impairs Both Primary and Secondary RIG-I Signaling for Interferon Induction in Human Monocytes. Sci. Signal. 2017, 10, eaan2392. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, T.S.; de Sá, K.S.G.; Ishimoto, A.Y.; Becerra, A.; Oliveira, S.; Almeida, L.; Gonçalves, A.V.; Perucello, D.B.; Andrade, W.A.; Castro, R.; et al. Inflammasomes Are Activated in Response to SARS-CoV-2 Infection and Are Associated with COVID-19 Severity in Patients. J. Exp. Med. 2021, 218, e20201707. [Google Scholar] [CrossRef]
- McGonagle, D.; Sharif, K.; O’Regan, A.; Bridgewood, C. The Role of Cytokines Including Interleukin-6 in COVID-19 Induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun. Rev. 2020, 19, 102537. [Google Scholar] [CrossRef]
- Sayed, N.; Huang, Y.; Nguyen, K.; Krejciova-Rajaniemi, Z.; Grawe, A.P.; Gao, T.; Tibshirani, R.; Hastie, T.; Alpert, A.; Cui, L.; et al. An Inflammatory Aging Clock (IAge) Based on Deep Learning Tracks Multimorbidity, Immunosenescence, Frailty and Cardiovascular Aging. Nat. Aging 2021, 1, 598–615. [Google Scholar] [CrossRef]
- Schmitt, C.A.; Tchkonia, T.; Niedernhofer, L.J.; Robbins, P.D.; Kirkland, J.L.; Lee, S. COVID-19 and Cellular Senescence. Nat. Rev. Immunol. 2022, 1–13. [Google Scholar] [CrossRef]
- Bartleson, J.M.; Radenkovic, D.; Covarrubias, A.J.; Furman, D.; Winer, D.A.; Verdin, E. SARS-CoV-2, COVID-19 and the Aging Immune System. Nat. Aging 2021, 1, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Bergsma, T.; Rogaeva, E. DNA Methylation Clocks and Their Predictive Capacity for Aging Phenotypes and Healthspan. Neurosci. Insights 2020, 15, 263310552094222. [Google Scholar] [CrossRef] [PubMed]
- McCrory, C.; Fiorito, G.; Hernandez, B.; Polidoro, S.; O’Halloran, A.M.; Hever, A.; Ni Cheallaigh, C.; Lu, A.T.; Horvath, S.; Vineis, P.; et al. GrimAge Outperforms Other Epigenetic Clocks in the Prediction of Age-Related Clinical Phenotypes and All-Cause Mortality. J. Gerontol. Ser. A 2021, 76, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Mongelli, A.; Barbi, V.; Gottardi Zamperla, M.; Atlante, S.; Forleo, L.; Nesta, M.; Massetti, M.; Pontecorvi, A.; Nanni, S.; Farsetti, A.; et al. Evidence for Biological Age Acceleration and Telomere Shortening in COVID-19 Survivors. Int. J. Mol. Sci. 2021, 22, 6151. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Codd, V.; Raisi-Estabragh, Z.; Musicha, C.; Bountziouka, V.; Kaptoge, S.; Allara, E.; Di Angelantonio, E.; Butterworth, A.S.; Wood, A.M.; et al. Shorter Leukocyte Telomere Length Is Associated with Adverse COVID-19 Outcomes: A Cohort Study in UK Biobank. EBioMedicine 2021, 70, 103485. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, W.; Wang, T.; Ran, D.; Davalos, V.; Planas-Serra, L.; Pujol, A.; Esteller, M.; Wang, X.; Yu, H. Accelerated Biological Aging in COVID-19 Patients. Nat. Commun. 2022, 13, 2135. [Google Scholar] [CrossRef]
- Bohlin, J.; Page, C.M.; Lee, Y.; Pettersson, J.H.-O.; Jugessur, A.; Magnus, P.; Håberg, S.E. Age and Sex Effects on DNA Methylation Sites Linked to Genes Implicated in Severe COVID-19 and SARS-CoV-2 Host Cell Entry. PLoS ONE 2022, 17, e0269105. [Google Scholar] [CrossRef]
- Corley, M.J.; Pang, A.P.S.; Dody, K.; Mudd, P.A.; Patterson, B.K.; Seethamraju, H.; Bram, Y.; Peluso, M.J.; Torres, L.; Iyer, N.S.; et al. Genome-wide DNA Methylation Profiling of Peripheral Blood Reveals an Epigenetic Signature Associated with Severe COVID-19. J. Leukoc. Biol. 2021, 110, 21–26. [Google Scholar] [CrossRef]
- Balnis, J.; Madrid, A.; Hogan, K.J.; Drake, L.A.; Chieng, H.C.; Tiwari, A.; Vincent, C.E.; Chopra, A.; Vincent, P.A.; Robek, M.D.; et al. Blood DNA Methylation and COVID-19 Outcomes. Clin. Epigenetics 2021, 13, 118. [Google Scholar] [CrossRef]
- Pang, A.P.S.; Higgins-Chen, A.T.; Comite, F.; Raica, I.; Arboleda, C.; Went, H.; Mendez, T.; Schotsaert, M.; Dwaraka, V.; Smith, R.; et al. Longitudinal Study of DNA Methylation and Epigenetic Clocks Prior to and Following Test-Confirmed COVID-19 and MRNA Vaccination. Front. Genet. 2022, 13, 819749. [Google Scholar] [CrossRef]
- Attia, M.H. A Cautionary Note on Altered Pace of Aging in the COVID-19 Era. Forensic Sci. Int. Genet. 2022, 59, 102724. [Google Scholar] [CrossRef] [PubMed]
- Kenmoe, S.; Kengne-Nde, C.; Ebogo-Belobo, J.T.; Mbaga, D.S.; Modiyinji, A.F.; Njouom, R. Systematic Review and Meta-Analysis of the Prevalence of Common Respiratory Viruses in Children < 2 Years with Bronchiolitis in the Pre-COVID-19 Pandemic Era. PLoS ONE 2020, 15, e0242302. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; O’Brien, K.L.; Madhi, S.A.; Widdowson, M.-A.; Byass, P.; Omer, S.B.; Abbas, Q.; Ali, A.; Amu, A.; et al. Global Burden of Respiratory Infections Associated with Seasonal Influenza in Children under 5 Years in 2018: A Systematic Review and Modelling Study. Lancet Glob. Health 2020, 8, e497–e510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, V.; Coleman, S.; Nixon, J.; Sharples, L.; Hamilton-Shield, J.; Rutter, H.; Bryant, M. A Systematic Review and Meta-Analysis Estimating the Population Prevalence of Comorbidities in Children and Adolescents Aged 5 to 18 Years. Obes. Rev. 2019, 20, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Pierce, C.A.; Preston-Hurlburt, P.; Dai, Y.; Aschner, C.B.; Cheshenko, N.; Galen, B.; Garforth, S.J.; Herrera, N.G.; Jangra, R.K.; Morano, N.C.; et al. Immune Responses to SARS-CoV-2 Infection in Hospitalized Pediatric and Adult Patients. Sci. Transl. Med. 2020, 12, eabd5487. [Google Scholar] [CrossRef]
- Pierce, C.A.; Sy, S.; Galen, B.; Goldstein, D.Y.; Orner, E.; Keller, M.J.; Herold, K.C.; Herold, B.C. Natural Mucosal Barriers and COVID-19 in Children. JCI Insight 2021, 6, e148694. [Google Scholar] [CrossRef]
- Yoshida, M.; Worlock, K.B.; Huang, N.; Lindeboom, R.G.H.; Butler, C.R.; Kumasaka, N.; Dominguez Conde, C.; Mamanova, L.; Bolt, L.; Richardson, L.; et al. Local and Systemic Responses to SARS-CoV-2 Infection in Children and Adults. Nature 2022, 602, 321–327. [Google Scholar] [CrossRef]
- Chou, J.; Thomas, P.G.; Randolph, A.G. Immunology of SARS-CoV-2 Infection in Children. Nat. Immunol. 2022, 23, 177–185. [Google Scholar] [CrossRef]
- Cohen, C.A.; Li, A.P.Y.; Hachim, A.; Hui, D.S.C.; Kwan, M.Y.W.; Tsang, O.T.Y.; Chiu, S.S.; Chan, W.H.; Yau, Y.S.; Kavian, N.; et al. SARS-CoV-2 Specific T Cell Responses Are Lower in Children and Increase with Age and Time after Infection. Nat. Commun. 2021, 12, 4678. [Google Scholar] [CrossRef]
- Vazquez, C.; Swanson, S.E.; Negatu, S.G.; Dittmar, M.; Miller, J.; Ramage, H.R.; Cherry, S.; Jurado, K.A. SARS-CoV-2 Viral Proteins NSP1 and NSP13 Inhibit Interferon Activation through Distinct Mechanisms. PLoS ONE 2021, 16, e0253089. [Google Scholar] [CrossRef]
- Loske, J.; Röhmel, J.; Lukassen, S.; Stricker, S.; Magalhães, V.G.; Liebig, J.; Chua, R.L.; Thürmann, L.; Messingschlager, M.; Seegebarth, A.; et al. Pre-Activated Antiviral Innate Immunity in the Upper Airways Controls Early SARS-CoV-2 Infection in Children. Nat. Biotechnol. 2022, 40, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Bauer, V.L.; Sawyer, S.L.; Diaz-Griffero, F. Human ACE2 Polymorphisms from Different Human Populations Modulate SARS-CoV-2 Infection. Viruses 2022, 14, 1451. [Google Scholar] [CrossRef]
- Dieter, C.; Brondani, L.d.A.; Leitão, C.B.; Gerchman, F.; Lemos, N.E.; Crispim, D. Genetic Polymorphisms Associated with Susceptibility to COVID-19 Disease and Severity: A Systematic Review and Meta-Analysis. PLoS ONE 2022, 17, e0270627. [Google Scholar] [CrossRef] [PubMed]
- Saengsiwaritt, W.; Jittikoon, J.; Chaikledkaew, U.; Udomsinprasert, W. Genetic Polymorphisms of ACE1, ACE2, and TMPRSS2 Associated with COVID-19 Severity: A Systematic Review with Meta-Analysis. Rev. Med. Virol. 2022, 32, e2323. [Google Scholar] [CrossRef]
- Russo, R.; Andolfo, I.; Lasorsa, V.A.; Iolascon, A.; Capasso, M. Genetic Analysis of the Coronavirus SARS-CoV-2 Host Protease TMPRSS2 in Different Populations. Front. Genet. 2020, 11, 872. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.E.K.; Daly, M.J.; Ganna, A. The Human Genetic Epidemiology of COVID-19. Nat. Rev. Genet. 2022, 23, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Pathak, G.A.; Karjalainen, J.; Stevens, C.; Neale, B.M.; Daly, M.; Ganna, A.; Andrews, S.J.; Kanai, M.; Cordioli, M.; Polimanti, R.; et al. A First Update on Mapping the Human Genetic Architecture of COVID-19. Nature 2022, 608, E1–E10. [Google Scholar] [CrossRef]
- van Moorsel, C.H.M.; van der Vis, J.J.; Duckworth, A.; Scotton, C.J.; Benschop, C.; Ellinghaus, D.; Ruven, H.J.T.; Quanjel, M.J.R.; Grutters, J.C. The MUC5B Promoter Polymorphism Associates with Severe COVID-19 in the European Population. Front. Med. 2021, 8, 668024. [Google Scholar] [CrossRef]
- Roberts, G.H.L.; Partha, R.; Rhead, B.; Knight, S.C.; Park, D.S.; Coignet, M.V.; Zhang, M.; Berkowitz, N.; Turrisini, D.A.; Gaddis, M.; et al. Expanded COVID-19 Phenotype Definitions Reveal Distinct Patterns of Genetic Association and Protective Effects. Nat. Genet. 2022, 54, 374–381. [Google Scholar] [CrossRef]
- Karlsen, T.H. Understanding COVID-19 through Genome-Wide Association Studies. Nat. Genet. 2022, 54, 368–369. [Google Scholar] [CrossRef]
- Downes, D.J.; Cross, A.R.; Hua, P.; Roberts, N.; Schwessinger, R.; Cutler, A.J.; Munis, A.M.; Brown, J.; Mielczarek, O.; de Andrea, C.E.; et al. Identification of LZTFL1 as a Candidate Effector Gene at a COVID-19 Risk Locus. Nat. Genet. 2021, 53, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.C.; Schmidt, A.G.; Bölke, E.; Uhrberg, M.; Keitel, V.; Feldt, T.; Jensen, B.; Häussinger, D.; Adams, O.; Schneider, E.M.; et al. Association of HLA Genotypes, AB0 Blood Type and Chemokine Receptor 5 Mutant CD195 with the Clinical Course of COVID-19. Eur. J. Med. Res. 2021, 26, 107. [Google Scholar] [CrossRef] [PubMed]
- Wickenhagen, A.; Sugrue, E.; Lytras, S.; Kuchi, S.; Noerenberg, M.; Turnbull, M.L.; Loney, C.; Herder, V.; Allan, J.; Jarmson, I.; et al. A Prenylated DsRNA Sensor Protects against Severe COVID-19. Science 2021, 374, eabj3624. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Butler-Laporte, G.; Nakanishi, T.; Morrison, D.R.; Afilalo, J.; Afilalo, M.; Laurent, L.; Pietzner, M.; Kerrison, N.; Zhao, K.; et al. A Neanderthal OAS1 Isoform Protects Individuals of European Ancestry against COVID-19 Susceptibility and Severity. Nat. Med. 2021, 27, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Boisson, B.; Onodi, F.; Matuozzo, D.; Moncada-Velez, M.; Maglorius Renkilaraj, M.R.L.; Zhang, P.; Meertens, L.; Bolze, A.; Materna, M.; et al. X-Linked Recessive TLR7 Deficiency in ~1% of Men under 60 Years Old with Life-Threatening COVID-19. Sci. Immunol. 2021, 6, eabl4348. [Google Scholar] [CrossRef]
- Namkoong, H.; Edahiro, R.; Takano, T.; Nishihara, H.; Shirai, Y.; Sonehara, K.; Tanaka, H.; Azekawa, S.; Mikami, Y.; Lee, H.; et al. DOCK2 Is Involved in the Host Genetics and Biology of Severe COVID-19. Nature 2022, 609, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Abbas, M.; Verma, S.; Khan, F.H.; Raza, S.T.; Siddiqi, Z.; Ahmad, I.; Mahdi, F. Impact of I/D Polymorphism of Angiotensin-Converting Enzyme 1 (ACE1) Gene on the Severity of COVID-19 Patients. Infect. Genet. Evol. 2021, 91, 104801. [Google Scholar] [CrossRef]
- Pietzner, M.; Chua, R.L.; Wheeler, E.; Jechow, K.; Willett, J.D.S.; Radbruch, H.; Trump, S.; Heidecker, B.; Zeberg, H.; Heppner, F.L.; et al. ELF5 Is a Potential Respiratory Epithelial Cell-Specific Risk Gene for Severe COVID-19. Nat. Commun. 2022, 13, 4484. [Google Scholar] [CrossRef]
- Hernández Cordero, A.I.; Li, X.; Milne, S.; Yang, C.X.; Bossé, Y.; Joubert, P.; Timens, W.; van den Berge, M.; Nickle, D.; Hao, K.; et al. Multi-Omics Highlights ABO Plasma Protein as a Causal Risk Factor for COVID-19. Hum. Genet. 2021, 140, 969–979. [Google Scholar] [CrossRef]
- Kurki, S.N.; Kantonen, J.; Kaivola, K.; Hokkanen, L.; Mäyränpää, M.I.; Puttonen, H.; Martola, J.; Pöyhönen, M.; Kero, M.; Tuimala, J.; et al. APOE Ε4 Associates with Increased Risk of Severe COVID-19, Cerebral Microhaemorrhages and Post-COVID Mental Fatigue: A Finnish Biobank, Autopsy and Clinical Study. Acta Neuropathol. Commun. 2021, 9, 199. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 13 July 2022).
- COVID-19 Host Genetics Initiative; Gazon, H.; Juszczak, D.; Fadeur, M.; Camby, S.; Meuris, C.; Thys, M.; Jacques, J.; Henket, M.; Beguin, Y.; et al. Mapping the Human Genetic Architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef]
- Huang, Q.-M.; Zhang, P.-D.; Li, Z.-H.; Zhou, J.-M.; Liu, D.; Zhang, X.-R.; Zhong, W.-F.; Zhang, Y.-J.; Shen, D.; Liang, F.; et al. Genetic Risk and Chronic Obstructive Pulmonary Disease Independently Predict the Risk of Incident Severe COVID-19. Ann. Am. Thorac. Soc. 2022, 19, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Fadista, J.; Manning, A.K.; Florez, J.C.; Groop, L. The (in)Famous GWAS P-Value Threshold Revisited and Updated for Low-Frequency Variants. Eur. J. Hum. Genet. 2016, 24, 1202–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tangye, S.G.; Al-Herz, W.; Bousfiha, A.; Chatila, T.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L.; Holland, S.M.; Klein, C.; Morio, T.; et al. Human Inborn Errors of Immunity: 2019 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2020, 40, 24–64. [Google Scholar] [CrossRef] [Green Version]
- van der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; van den Heuvel, G.; Mantere, T.; Kersten, S.; van Deuren, R.C.; Steehouwer, M.; van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of Genetic Variants among Young Men with Severe COVID-19. JAMA 2020, 324, 663–673. [Google Scholar] [CrossRef]
- Fallerini, C.; Daga, S.; Mantovani, S.; Benetti, E.; Picchiotti, N.; Francisci, D.; Paciosi, F.; Schiaroli, E.; Baldassarri, M.; Fava, F.; et al. Association of Toll-like Receptor 7 Variants with Life-Threatening COVID-19 Disease in Males: Findings from a Nested Case-Control Study. eLife 2021, 10, e67569. [Google Scholar] [CrossRef]
- Mantovani, S.; Daga, S.; Fallerini, C.; Baldassarri, M.; Benetti, E.; Picchiotti, N.; Fava, F.; Gallì, A.; Zibellini, S.; Bruttini, M.; et al. Rare Variants in Toll-like Receptor 7 Results in Functional Impairment and Downregulation of Cytokine-Mediated Signaling in COVID-19 Patients. Genes Immun. 2022, 23, 51–56. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- Kosmicki, J.A.; Horowitz, J.E.; Banerjee, N.; Lanche, R.; Marcketta, A.; Maxwell, E.; Bai, X.; Sun, D.; Backman, J.D.; Sharma, D.; et al. Pan-Ancestry Exome-Wide Association Analyses of COVID-19 Outcomes in 586,157 Individuals. Am. J. Hum. Genet. 2021, 108, 1350–1355. [Google Scholar] [CrossRef]
- Povysil, G.; Butler-Laporte, G.; Shang, N.; Wang, C.; Khan, A.; Alaamery, M.; Nakanishi, T.; Zhou, S.; Forgetta, V.; Eveleigh, R.J.M.; et al. Rare Loss-of-Function Variants in Type I IFN Immunity Genes Are Not Associated with Severe COVID-19. J. Clin. Investig. 2021, 131, e147834. [Google Scholar] [CrossRef]
- Drzymalla, E.; Green, R.F.; Knuth, M.; Khoury, M.J.; Dotson, W.D.; Gundlapalli, A. COVID-19-Related Health Outcomes in People with Primary Immunodeficiency: A Systematic Review. Clin. Immunol. 2022, 243, 109097. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Gervais, A.; Le Voyer, T.; Rosain, J.; Philippot, Q.; Manry, J.; Michailidis, E.; Hoffmann, H.-H.; Eto, S.; Garcia-Prat, M.; et al. Autoantibodies Neutralizing Type I IFNs Are Present in ~4% of Uninfected Individuals over 70 Years Old and Account for ~20% of COVID-19 Deaths. Sci. Immunol. 2021, 6, eabl4340. [Google Scholar] [CrossRef] [PubMed]
- Briquez, P.S.; Rouhani, S.J.; Yu, J.; Pyzer, A.R.; Trujillo, J.; Dugan, H.L.; Stamper, C.T.; Changrob, S.; Sperling, A.I.; Wilson, P.C.; et al. Severe COVID-19 Induces Autoantibodies against Angiotensin II That Correlate with Blood Pressure Dysregulation and Disease Severity. Sci. Adv. 2022, 8, eabn3777. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Domínguez-Andrés, J.; van de Veerdonk, F.L.; van Crevel, R.; Pulendran, B.; van der Meer, J.W.M. Natural Resistance against Infections: Focus on COVID-19. Trends Immunol. 2022, 43, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Andreakos, E.; Abel, L.; Vinh, D.C.; Kaja, E.; Drolet, B.A.; Zhang, Q.; O’Farrelly, C.; Novelli, G.; Rodríguez-Gallego, C.; Haerynck, F.; et al. A Global Effort to Dissect the Human Genetic Basis of Resistance to SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 159–164. [Google Scholar] [CrossRef]
- David, A.; Parkinson, N.; Peacock, T.P.; Pairo-Castineira, E.; Khanna, T.; Cobat, A.; Tenesa, A.; Sancho-Shimizu, V.; Casanova, J.-L.; Abel, L.; et al. A Common TMPRSS2 Variant Has a Protective Effect against Severe COVID-19. Curr. Res. Transl. Med. 2022, 70, 103333. [Google Scholar] [CrossRef]
- Verma, A.; Minnier, J.; Wan, E.S.; Huffman, J.E.; Gao, L.; Joseph, J.; Ho, Y.-L.; Wu, W.-C.; Cho, K.; Gorman, B.R.; et al. A MUC5B Gene Polymorphism, Rs35705950-T Confers Protective Effects Against COVID-19 Hospitalization but Not Severe Disease or Mortality. Am. J. Respir. Crit. Care Med. 2022, 206, 1220–1229. [Google Scholar] [CrossRef]
- Shkurnikov, M.; Nersisyan, S.; Jankevic, T.; Galatenko, A.; Gordeev, I.; Vechorko, V.; Tonevitsky, A. Association of HLA Class I Genotypes with Severity of Coronavirus Disease-19. Front. Immunol. 2021, 12, 641900. [Google Scholar] [CrossRef]
- Hernández-Doño, S.; Sánchez-González, R.A.; Trujillo-Vizuet, M.G.; Zamudio-Castellanos, F.Y.; García-Silva, R.; Bulos-Rodríguez, P.; Vazquez-Guzmán, C.A.; Cárdenas-Ramos, X.; de León Rodríguez, D.; Elías, F.; et al. Protective HLA Alleles against Severe COVID-19: HLA-A*68 as an Ancestral Protection Allele in Tapachula-Chiapas, Mexico. Clin. Immunol. 2022, 238, 108990. [Google Scholar] [CrossRef]
- Abdelhafiz, A.S.; Ali, A.; Fouda, M.A.; Sayed, D.M.; Kamel, M.M.; Kamal, L.M.; Khalil, M.A.; Bakry, R.M. HLA-B*15 Predicts Survival in Egyptian Patients with COVID-19. Hum. Immunol. 2022, 83, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.; Felipe, S.; de Freitas, R.; Araújo, V.; Soares, P.; Ribeiro, J.; Henrique dos Santos, L.; Alves, J.O.; Canabrava, N.; van Tilburg, M.; et al. ABO Blood Group and Link to COVID-19: A Comprehensive Review of the Reported Associations and Their Possible Underlying Mechanisms. Microb. Pathog. 2022, 169, 105658. [Google Scholar] [CrossRef] [PubMed]
- Magesh, S.; John, D.; Li, W.T.; Li, Y.; Mattingly-app, A.; Jain, S.; Chang, E.Y.; Ongkeko, W.M. Disparities in COVID-19 Outcomes by Race, Ethnicity, and Socioeconomic Status: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2021, 4, e2134147. [Google Scholar] [CrossRef] [PubMed]
- Parcha, V.; Malla, G.; Suri, S.S.; Kalra, R.; Heindl, B.; Berra, L.; Fouad, M.N.; Arora, G.; Arora, P. Geographic Variation in Racial Disparities in Health and Coronavirus Disease-2019 (COVID-19) Mortality. Mayo Clin. Proc. Innov. Qual. Outcomes 2020, 4, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Price, L.S.; Nattinger, A.B.; Rivera, F.; Hanson, R.; Gmehlin, C.G.; Perez, A.; Singh, S.; Buchan, B.W.; Ledeboer, N.A.; Pezzin, L.E. Racial Disparities in Incidence and Outcomes among Patients with COVID-19. JAMA Netw. Open 2020, 3, e2021892. [Google Scholar] [CrossRef] [PubMed]
- Shelton, J.F.; Shastri, A.J.; Ye, C.; Weldon, C.H.; Filshtein-Sonmez, T.; Coker, D.; Symons, A.; Esparza-Gordillo, J.; Aslibekyan, S.; Auton, A. Trans-Ancestry Analysis Reveals Genetic and Nongenetic Associations with COVID-19 Susceptibility and Severity. Nat. Genet. 2021, 53, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Raharja, A.; Tamara, A.; Kok, L.T. Association Between Ethnicity and Severe COVID-19 Disease: A Systematic Review and Meta-Analysis. J. Racial Ethn. Health Disparities 2021, 8, 1563–1572. [Google Scholar] [CrossRef]
- Li, S.L.; Pereira, R.H.; Prete, C.A., Jr.; Zarebski, A.E.; Emanuel, L.; Alves, P.J.; Peixoto, P.S.; Braga, C.K.; de Souza Santos, A.A.; de Souza, W.M.; et al. Higher Risk of Death from COVID-19 in Low-Income and Non-White Populations of São Paulo, Brazil. BMJ Glob. Health 2021, 6, e004959. [Google Scholar] [CrossRef]
- Zeberg, H.; Pääbo, S. The Major Genetic Risk Factor for Severe COVID-19 Is Inherited from Neanderthals. Nature 2020, 587, 610–612. [Google Scholar] [CrossRef]
- Zeberg, H.; Pääbo, S. A Genomic Region Associated with Protection against Severe COVID-19 Is Inherited from Neandertals. Proc. Natl. Acad. Sci. USA 2021, 118, e2026309118. [Google Scholar] [CrossRef]
- Falagas, M.E.; Mourtzoukou, E.G.; Vardakas, K.Z. Sex Differences in the Incidence and Severity of Respiratory Tract Infections. Respir. Med. 2007, 101, 1845–1863. [Google Scholar] [CrossRef]
- Ursin, R.L.; Klein, S.L. Sex Differences in Respiratory Viral Pathogenesis and Treatments. Annu. Rev. Virol. 2021, 8, 393–414. [Google Scholar] [CrossRef] [PubMed]
- Corica, B.; Tartaglia, F.; D’Amico, T.; Romiti, G.F.; Cangemi, R. Sex and Gender Differences in Community-Acquired Pneumonia. Intern. Emerg. Med. 2022, 17, 1575–1588. [Google Scholar] [CrossRef]
- Sieurin, J.; Brandén, G.; Magnusson, C.; Hergens, M.-P.; Kosidou, K. A Population-Based Cohort Study of Sex and Risk of Severe Outcomes in COVID-19. Eur. J. Epidemiol. 2022, 37, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Meijs, D.A.M.; van Bussel, B.C.T.; Stessel, B.; Mehagnoul-Schipper, J.; Hana, A.; Scheeren, C.I.E.; Peters, S.A.E.; van Mook, W.N.K.A.; van der Horst, I.C.C.; Marx, G.; et al. Better COVID-19 Intensive Care Unit Survival in Females, Independent of Age, Disease Severity, Comorbidities, and Treatment. Sci. Rep. 2022, 12, 734. [Google Scholar] [CrossRef]
- Her, A.-Y.; Bhak, Y.; Jun, E.J.; Yuan, S.L.; Garg, S.; Lee, S.; Bhak, J.; Shin, E.-S. Sex-Specific Difference of in-Hospital Mortality from COVID-19 in South Korea. PLoS ONE 2022, 17, e0262861. [Google Scholar] [CrossRef] [PubMed]
- Vahidy, F.S.; Pan, A.P.; Ahnstedt, H.; Munshi, Y.; Choi, H.A.; Tiruneh, Y.; Nasir, K.; Kash, B.A.; Andrieni, J.D.; McCullough, L.D. Sex Differences in Susceptibility, Severity, and Outcomes of Coronavirus Disease 2019: Cross-Sectional Analysis from a Diverse US Metropolitan Area. PLoS ONE 2021, 16, e0245556. [Google Scholar] [CrossRef]
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male Sex Identified by Global COVID-19 Meta-Analysis as a Risk Factor for Death and ITU Admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef]
- Bennett, T.D.; Moffitt, R.A.; Hajagos, J.G.; Amor, B.; Anand, A.; Bissell, M.M.; Bradwell, K.R.; Bremer, C.; Byrd, J.B.; Denham, A.; et al. Clinical Characterization and Prediction of Clinical Severity of SARS-CoV-2 Infection among US Adults Using Data from the US National COVID Cohort Collaborative. JAMA Netw. Open 2021, 4, e2116901. [Google Scholar] [CrossRef]
- Gujski, M.; Jankowski, M.; Rabczenko, D.; Goryński, P.; Juszczyk, G. Characteristics and Clinical Outcomes of 116,539 Patients Hospitalized with COVID-19—Poland, March–December 2020. Viruses 2021, 13, 1458. [Google Scholar] [CrossRef]
- Kim, H.-J.; Hwang, H.; Hong, H.; Yim, J.-J.; Lee, J. A Systematic Review and Meta-Analysis of Regional Risk Factors for Critical Outcomes of COVID-19 during Early Phase of the Pandemic. Sci. Rep. 2021, 11, 9784. [Google Scholar] [CrossRef]
- Dessie, Z.G.; Zewotir, T. Mortality-Related Risk Factors of COVID-19: A Systematic Review and Meta-Analysis of 42 Studies and 423,117 Patients. BMC Infect. Dis. 2021, 21, 855. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhong, X.; Wang, Y.; Zeng, X.; Luo, T.; Liu, Q. Clinical Determinants of the Severity of COVID-19: A Systematic Review and Meta-Analysis. PLoS ONE 2021, 16, e0250602. [Google Scholar] [CrossRef]
- Rivero-Calle, I.; Pardo-Seco, J.; Aldaz, P.; Vargas, D.A.; Mascarós, E.; Redondo, E.; Díaz-Maroto, J.L.; Linares-Rufo, M.; Fierro-Alacio, M.J.; Gil, A.; et al. Incidence and Risk Factor Prevalence of Community-Acquired Pneumonia in Adults in Primary Care in Spain (NEUMO-ES-RISK Project). BMC Infect. Dis. 2016, 16, 645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higgins, S.T.; Kurti, A.N.; Redner, R.; White, T.J.; Gaalema, D.E.; Roberts, M.E.; Doogan, N.J.; Tidey, J.W.; Miller, M.E.; Stanton, C.A.; et al. A Literature Review on Prevalence of Gender Differences and Intersections with Other Vulnerabilities to Tobacco Use in the United States, 2004–2014. Prev. Med. 2015, 80, 89–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamekh, M.; Deny, M.; Romano, M.; Lefèvre, N.; Corazza, F.; Duchateau, J.; Casimir, G. Differential Susceptibility to Infectious Respiratory Diseases between Males and Females Linked to Sex-Specific Innate Immune Inflammatory Response. Front. Immunol. 2017, 8, 1806. [Google Scholar] [CrossRef] [PubMed]
- Kadel, S.; Kovats, S. Sex Hormones Regulate Innate Immune Cells and Promote Sex Differences in Respiratory Virus Infection. Front. Immunol. 2018, 9, 1653. [Google Scholar] [CrossRef] [Green Version]
- Shepherd, R.; Cheung, A.S.; Pang, K.; Saffery, R.; Novakovic, B. Sexual Dimorphism in Innate Immunity: The Role of Sex Hormones and Epigenetics. Front. Immunol. 2021, 11, 604000. [Google Scholar] [CrossRef]
- Brandi, M.L. Are Sex Hormones Promising Candidates to Explain Sex Disparities in the COVID-19 Pandemic? Rev. Endocr. Metab. Disord. 2022, 23, 171–183. [Google Scholar] [CrossRef]
- Gubbels Bupp, M.R.; Jorgensen, T.N. Androgen-Induced Immunosuppression. Front. Immunol. 2018, 9, 794. [Google Scholar] [CrossRef] [Green Version]
- Decaroli, M.C.; Rochira, V. Aging and Sex Hormones in Males. Virulence 2017, 8, 545–570. [Google Scholar] [CrossRef]
- Rastrelli, G.; Di Stasi, V.; Inglese, F.; Beccaria, M.; Garuti, M.; Di Costanzo, D.; Spreafico, F.; Greco, G.F.; Cervi, G.; Pecoriello, A.; et al. Low Testosterone Levels Predict Clinical Adverse Outcomes in SARS-CoV-2 Pneumonia Patients. Andrology 2021, 9, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Pagano, M.T.; Peruzzu, D.; Busani, L.; Pierdominici, M.; Ruggieri, A.; Antinori, A.; D’Offizi, G.; Petrosillo, N.; Palmieri, F.; Piselli, P.; et al. Predicting Respiratory Failure in Patients Infected by SARS-CoV-2 by Admission Sex-Specific Biomarkers. Biol. Sex Differ. 2021, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Baldassarri, M.; Picchiotti, N.; Fava, F.; Fallerini, C.; Benetti, E.; Daga, S.; Valentino, F.; Doddato, G.; Furini, S.; Giliberti, A.; et al. Shorter Androgen Receptor PolyQ Alleles Protect against Life-Threatening COVID-19 Disease in European Males. EBioMedicine 2021, 65, 103246. [Google Scholar] [CrossRef]
- Montaño, L.M.; Sommer, B.; Solís-Chagoyán, H.; Romero-Martínez, B.S.; Aquino-Gálvez, A.; Gomez-Verjan, J.C.; Calixto, E.; González-Avila, G.; Flores-Soto, E. Could Lower Testosterone in Older Men Explain Higher COVID-19 Morbidity and Mortalities? Int. J. Mol. Sci. 2022, 23, 935. [Google Scholar] [CrossRef] [PubMed]
- Traish, A.; Bolanos, J.; Nair, S.; Saad, F.; Morgentaler, A. Do Androgens Modulate the Pathophysiological Pathways of Inflammation? Appraising the Contemporary Evidence. J. Clin. Med. 2018, 7, 549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, B.; Ferguson, C.; White, J.T.; Wang, S.; Vessella, R.; True, L.D.; Hood, L.; Nelson, P.S. Prostate-Localized and Androgen-Regulated Expression of the Membrane-Bound Serine Protease TMPRSS2. Cancer Res. 1999, 59, 4180–4184. [Google Scholar]
- Wang, X.-W.; Hu, H.; Xu, Z.-Y.; Zhang, G.-K.; Yu, Q.-H.; Yang, H.-L.; Zheng, J.-H. Association of Menopausal Status with COVID-19 Outcomes: A Propensity Score Matching Analysis. Biol. Sex Differ. 2021, 12, 16. [Google Scholar] [CrossRef]
- Sund, M.; Fonseca-Rodríguez, O.; Josefsson, A.; Welen, K.; Connolly, A.-M.F. Association between Pharmaceutical Modulation of Oestrogen in Postmenopausal Women in Sweden and Death Due to COVID-19: A Cohort Study. BMJ Open 2022, 12, e053032. [Google Scholar] [CrossRef]
- Costeira, R.; Lee, K.A.; Murray, B.; Christiansen, C.; Castillo-Fernandez, J.; Lochlainn, M.N.; Pujol, J.C.; Macfarlane, H.; Kenny, L.C.; Buchan, I.; et al. Estrogen and COVID-19 Symptoms: Associations in Women from the COVID Symptom Study. PLoS ONE 2021, 16, e0257051. [Google Scholar] [CrossRef]
- Mompeón, A.; Lázaro-Franco, M.; Bueno-Betí, C.; Pérez-Cremades, D.; Vidal-Gómez, X.; Monsalve, E.; Gironacci, M.M.; Hermenegildo, C.; Novella, S. Estradiol, Acting through ERα, Induces Endothelial Non-Classic Renin-Angiotensin System Increasing Angiotensin 1-7 Production. Mol. Cell. Endocrinol. 2016, 422, 1–8. [Google Scholar] [CrossRef]
- Baristaite, G.; Gurwitz, D. Estradiol Reduces ACE2 and TMPRSS2 MRNA Levels in A549 Human Lung Epithelial Cells. Drug Dev. Res. 2022, 83, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Kovats, S. Estrogen Receptors Regulate Innate Immune Cells and Signaling Pathways. Cell. Immunol. 2015, 294, 63–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pernis, A.B. Estrogen and CD4+ T Cells. Curr. Opin. Rheumatol. 2007, 19, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Prossnitz, E.R. Mechanisms of Estradiol-Induced Insulin Secretion by the G Protein-Coupled Estrogen Receptor GPR30/GPER in Pancreatic β-Cells. Endocrinology 2011, 152, 3030–3039. [Google Scholar] [CrossRef] [PubMed]
- Iorga, A.; Cunningham, C.M.; Moazeni, S.; Ruffenach, G.; Umar, S.; Eghbali, M. The Protective Role of Estrogen and Estrogen Receptors in Cardiovascular Disease and the Controversial Use of Estrogen Therapy. Biol. Sex Differ. 2017, 8, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asselta, R.; Paraboschi, E.M.; Mantovani, A.; Duga, S. ACE2 and TMPRSS2 Variants and Expression as Candidates to Sex and Country Differences in COVID-19 Severity in Italy. Aging 2020, 12, 10087–10098. [Google Scholar] [CrossRef]
- Baughn, L.B.; Sharma, N.; Elhaik, E.; Sekulic, A.; Bryce, A.H.; Fonseca, R. Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clin. Proc. 2020, 95, 1989–1999. [Google Scholar] [CrossRef]
- Xie, X.; Xudong, X.; Chen, J.; Junzhu, C.; Wang, X.; Xingxiang, W.; Zhang, F.; Furong, Z.; Liu, Y.; Yanrong, L. Age- and Gender-Related Difference of ACE2 Expression in Rat Lung. Life Sci. 2006, 78, 2166–2171. [Google Scholar] [CrossRef]
- Okwan-Duodu, D.; Lim, E.-C.; You, S.; Engman, D.M. TMPRSS2 Activity May Mediate Sex Differences in COVID-19 Severity. Signal Transduct. Target. Ther. 2021, 6, 100. [Google Scholar] [CrossRef]
- Spiering, A.E.; de Vries, T.J. Why Females Do Better: The X Chromosomal TLR7 Gene-Dose Effect in COVID-19. Front. Immunol. 2021, 12, 756262. [Google Scholar] [CrossRef]
- Maan, A.A.; Eales, J.; Akbarov, A.; Rowland, J.; Xu, X.; Jobling, M.A.; Charchar, F.J.; Tomaszewski, M. The Y Chromosome: A Blueprint for Men’s Health? Eur. J. Hum. Genet. 2017, 25, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Dumanski, J.P.; Halvardson, J.; Davies, H.; Rychlicka-Buniowska, E.; Mattisson, J.; Moghadam, B.T.; Nagy, N.; Węglarczyk, K.; Bukowska-Strakova, K.; Danielsson, M.; et al. Immune Cells Lacking Y Chromosome Show Dysregulation of Autosomal Gene Expression. Cell. Mol. Life Sci. 2021, 78, 4019–4033. [Google Scholar] [CrossRef]
- Thompson, D.J.; Genovese, G.; Halvardson, J.; Ulirsch, J.C.; Wright, D.J.; Terao, C.; Davidsson, O.B.; Day, F.R.; Sulem, P.; Jiang, Y.; et al. Genetic Predisposition to Mosaic Y Chromosome Loss in Blood. Nature 2019, 575, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Flanagan, K.L. Sex Differences in Immune Responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.E.; Richardson, B.; Goncin, U.; McNeil, M.; Rioux, M.; Foley, M.K.; Ge, A.; Pechous, R.D.; Kindrachuk, J.; Cameron, C.M.; et al. Sex and Age Bias Viral Burden and Interferon Responses during SARS-CoV-2 Infection in Ferrets. Sci. Rep. 2021, 11, 14536. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; An, H.; Zhou, T.; Li, T.; Xie, M.; Chen, S.; Chen, C.; Ying, B.; Xu, Z.; Li, X.; et al. Sex- and Age-Specific Clinical and Immunological Features of Coronavirus Disease 2019. PLOS Pathog. 2021, 17, e1009420. [Google Scholar] [CrossRef]
- Takahashi, T.; Ellingson, M.K.; Wong, P.; Israelow, B.; Lucas, C.; Klein, J.; Silva, J.; Mao, T.; Oh, J.E.; Tokuyama, M.; et al. Sex Differences in Immune Responses That Underlie COVID-19 Disease Outcomes. Nature 2020, 588, 315–320. [Google Scholar] [CrossRef]
- Shattuck-Heidorn, H.; Danielsen, A.C.; Gompers, A.; Bruch, J.D.; Zhao, H.; Boulicault, M.; Marsella, J.; Richardson, S.S. A Finding of Sex Similarities Rather than Differences in COVID-19 Outcomes. Nature 2021, 597, E7–E9. [Google Scholar] [CrossRef]
- Lieberman, N.A.P.; Peddu, V.; Xie, H.; Shrestha, L.; Huang, M.-L.; Mears, M.C.; Cajimat, M.N.; Bente, D.A.; Shi, P.-Y.; Bovier, F.; et al. In Vivo Antiviral Host Transcriptional Response to SARS-CoV-2 by Viral Load, Sex, and Age. PLoS Biol. 2020, 18, e3000849. [Google Scholar] [CrossRef]
- Vassilaki, N.; Gargalionis, A.N.; Bletsa, A.; Papamichalopoulos, N.; Kontou, E.; Gkika, M.; Patas, K.; Theodoridis, D.; Manolis, I.; Ioannidis, A.; et al. Impact of Age and Sex on Antibody Response Following the Second Dose of COVID-19 BNT162b2 MRNA Vaccine in Greek Healthcare Workers. Microorganisms 2021, 9, 1725. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against Type I IFNs in Patients with Life-Threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ebinger, J.E.; Mostafa, R.; Budde, P.; Gajewski, J.; Walker, B.; Joung, S.; Wu, M.; Bräutigam, M.; Hesping, F.; et al. Paradoxical Sex-Specific Patterns of Autoantibody Response to SARS-CoV-2 Infection. J. Transl. Med. 2021, 19, 524. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.P.; Schumock, G.; Fu, M.; Massaccesi, G.; Muschelli, J.; Betz, J.; Klein, E.Y.; West, N.E.; Robinson, M.; Garibaldi, B.T.; et al. Sex and Gender Differences in Testing, Hospital Admission, Clinical Presentation, and Drivers of Severe Outcomes from COVID-19. Open Forum Infect. Dis. 2021, 8, ofab448. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, S.V.; Rusu, R.; Chan, B.; Bellows, M.; O’Keefe, C.; Nicholson, S. Sex Differences in Sequelae from COVID-19 Infection and in Long COVID Syndrome: A Review. Curr. Med. Res. Opin. 2022, 38, 1391–1399. [Google Scholar] [CrossRef]
- Bucciarelli, V.; Nasi, M.; Bianco, F.; Seferovic, J.; Ivkovic, V.; Gallina, S.; Mattioli, A.V. Depression Pandemic and Cardiovascular Risk in the COVID-19 Era and Long COVID Syndrome: Gender Makes a Difference. Trends Cardiovasc. Med. 2022, 32, 12–17. [Google Scholar] [CrossRef]
- Robinson, D.P.; Lorenzo, M.E.; Jian, W.; Klein, S.L. Elevated 17β-Estradiol Protects Females from Influenza A Virus Pathogenesis by Suppressing Inflammatory Responses. PLoS Pathog. 2011, 7, e1002149. [Google Scholar] [CrossRef] [Green Version]
- Di Mascio, D.; Khalil, A.; Saccone, G.; Rizzo, G.; Buca, D.; Liberati, M.; Vecchiet, J.; Nappi, L.; Scambia, G.; Berghella, V.; et al. Outcome of Coronavirus Spectrum Infections (SARS, MERS, COVID-19) during Pregnancy: A Systematic Review and Meta-Analysis. Am. J. Obstet. Gynecol. MFM 2020, 2, 100107. [Google Scholar] [CrossRef]
- Huntley, B.J.F.; Mulder, I.A.; Di Mascio, D.; Vintzileos, W.S.; Vintzileos, A.M.; Berghella, V.; Chauhan, S.P. Adverse Pregnancy Outcomes among Individuals with and without Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): A Systematic Review and Meta-Analysis. Obstet. Gynecol. 2021, 137, 585–596. [Google Scholar] [CrossRef]
- Gale, C.; Quigley, M.A.; Placzek, A.; Knight, M.; Ladhani, S.; Draper, E.S.; Sharkey, D.; Doherty, C.; Mactier, H.; Kurinczuk, J.J. Characteristics and Outcomes of Neonatal SARS-CoV-2 Infection in the UK: A Prospective National Cohort Study Using Active Surveillance. Lancet Child Adolesc. Health 2021, 5, 113–121. [Google Scholar] [CrossRef]
- Di Toro, F.; Gjoka, M.; Di Lorenzo, G.; De Santo, D.; De Seta, F.; Maso, G.; Risso, F.M.; Romano, F.; Wiesenfeld, U.; Levi-D’Ancona, R.; et al. Impact of COVID-19 on Maternal and Neonatal Outcomes: A Systematic Review and Meta-Analysis. Clin. Microbiol. Infect. 2021, 27, 36–46. [Google Scholar] [CrossRef]
- Martinez-Portilla, R.J.; Sotiriadis, A.; Chatzakis, C.; Torres-Torres, J.; Espino y Sosa, S.; Sandoval-Mandujano, K.; Castro-Bernabe, D.A.; Medina-Jimenez, V.; Monarrez-Martin, J.C.; Figueras, F.; et al. Pregnant Women with SARS-CoV-2 Infection Are at Higher Risk of Death and Pneumonia: Propensity Score Matched Analysis of a Nationwide Prospective Cohort (COV19Mx). Ultrasound Obstet. Gynecol. 2021, 57, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Allotey, J.; Stallings, E.; Bonet, M.; Yap, M.; Chatterjee, S.; Kew, T.; Debenham, L.; Llavall, A.C.; Dixit, A.; Zhou, D.; et al. Clinical Manifestations, Risk Factors, and Maternal and Perinatal Outcomes of Coronavirus Disease 2019 in Pregnancy: Living Systematic Review and Meta-Analysis. BMJ 2020, 370, m3320. [Google Scholar] [CrossRef] [PubMed]
- Rozo, N.; Valencia, D.; Newton, S.M.; Avila, G.; Gonzalez, M.A.; Sancken, C.L.; Burkel, V.K.; Ellington, S.R.; Gilboa, S.M.; Rao, C.Y.; et al. Severity of Illness by Pregnancy Status among Laboratory-Confirmed SARS-CoV-2 Infections Occurring in Reproductive-Aged Women in Colombia. Paediatr. Perinat. Epidemiol. 2022, 36, 456–465. [Google Scholar] [CrossRef]
- Marchand, G.; Patil, A.S.; Masoud, A.T.; Ware, K.; King, A.; Ruther, S.; Brazil, G.; Calteux, N.; Ulibarri, H.; Parise, J.; et al. Systematic Review and Meta-Analysis of COVID-19 Maternal and Neonatal Clinical Features and Pregnancy Outcomes up to June 3, 2021. AJOG Glob. Rep. 2022, 2, 100049. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.Q.; Bilodeau-Bertrand, M.; Liu, S.; Auger, N. The Impact of COVID-19 on Pregnancy Outcomes: A Systematic Review and Meta-Analysis. Can. Med. Assoc. J. 2021, 193, E540–E548. [Google Scholar] [CrossRef]
- Cruz Melguizo, S.; de la Cruz Conty, M.L.; Carmona Payán, P.; Abascal-Saiz, A.; Pintando Recarte, P.; González Rodríguez, L.; Cuenca Marín, C.; Martínez Varea, A.; Oreja Cuesta, A.B.; Rodríguez, P.P.; et al. Pregnancy Outcomes and SARS-CoV-2 Infection: The Spanish Obstetric Emergency Group Study. Viruses 2021, 13, 853. [Google Scholar] [CrossRef]
- Engjom, H.; Ramakrishnan, R.; Vousden, N.; Bunch, K.; Morris, E.; Simpson, N.A.B.; Gale, C.; O’Brien, P.; Quigley, M.; Brocklehurst, P.; et al. Severity of Maternal Infection and Perinatal Outcomes during Periods in Which Wildtype, Alpha and Delta SARS-CoV-2 Variants Were Dominant: Data from the UK Obstetric Surveillance System National Cohort Study. BMJ Med. 2022, 1, e000190. [Google Scholar] [CrossRef]
- Doyle, T.J.; Kiros, G.; Schmitt-Matzen, E.N.; Propper, R.; Thompson, A.; Phillips-Bell, G.S. Maternal and Perinatal Outcomes Associated with SARS-CoV-2 Infection during Pregnancy, Florida, 2020–2021: A Retrospective Cohort Study. Clin. Infect. Dis. 2022, 75, ciac441. [Google Scholar] [CrossRef]
- Ciapponi, A.; Bardach, A.; Comandé, D.; Berrueta, M.; Argento, F.J.; Rodriguez Cairoli, F.; Zamora, N.; Santa María, V.; Xiong, X.; Zaraa, S.; et al. COVID-19 and Pregnancy: An Umbrella Review of Clinical Presentation, Vertical Transmission, and Maternal and Perinatal Outcomes. PLoS ONE 2021, 16, e0253974. [Google Scholar] [CrossRef]
- Norman, M.; Navér, L.; Söderling, J.; Ahlberg, M.; Hervius Askling, H.; Aronsson, B.; Byström, E.; Jonsson, J.; Sengpiel, V.; Ludvigsson, J.F.; et al. Association of Maternal SARS-CoV-2 Infection in Pregnancy with Neonatal Outcomes. JAMA 2021, 325, 2076–2086. [Google Scholar] [CrossRef]
- Crovetto, F.; Crispi, F.; Llurba, E.; Pascal, R.; Larroya, M.; Trilla, C.; Camacho, M.; Medina, C.; Dobaño, C.; Gomez-Roig, M.D.; et al. Impact of Severe Acute Respiratory Syndrome Coronavirus 2 Infection on Pregnancy Outcomes: A Population-Based Study. Clin. Infect. Dis. 2021, 73, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Regan, A.K.; Arah, O.A.; Fell, D.B.; Sullivan, S.G. SARS-CoV-2 Infection During Pregnancy and Associated Perinatal Health Outcomes: A National US Cohort Study. J. Infect. Dis. 2022, 225, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Metz, T.D.; Clifton, R.G.; Hughes, B.L.; Sandoval, G.J.; Grobman, W.A.; Saade, G.R.; Manuck, T.A.; Longo, M.; Sowles, A.; Clark, K.; et al. Association of SARS-CoV-2 Infection with Serious Maternal Morbidity and Mortality from Obstetric Complications. JAMA 2022, 327, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Gurol-Urganci, I.; Jardine, J.E.; Carroll, F.; Draycott, T.; Dunn, G.; Fremeaux, A.; Harris, T.; Hawdon, J.; Morris, E.; Muller, P.; et al. Maternal and Perinatal Outcomes of Pregnant Women with SARS-CoV-2 Infection at the Time of Birth in England: National Cohort Study. Am. J. Obstet. Gynecol. 2021, 225, 522.e1–522.e11. [Google Scholar] [CrossRef] [PubMed]
- Piekos, S.N.; Roper, R.T.; Hwang, Y.M.; Sorensen, T.; Price, N.D.; Hood, L.; Hadlock, J.J. The Effect of Maternal SARS-CoV-2 Infection Timing on Birth Outcomes: A Retrospective Multicentre Cohort Study. Lancet Digit. Health 2022, 4, e95–e104. [Google Scholar] [CrossRef] [PubMed]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Luca, F.; Xu, Y.; Alazizi, A.; Leng, Y.; Hsu, C.-D.; Gomez-Lopez, N. Does the Human Placenta Express the Canonical Cell Entry Mediators for SARS-CoV-2? eLife 2020, 9, e58716. [Google Scholar] [CrossRef]
- Ashary, N.; Bhide, A.; Chakraborty, P.; Colaco, S.; Mishra, A.; Chhabria, K.; Jolly, M.K.; Modi, D. Single-Cell RNA-Seq Identifies Cell Subsets in Human Placenta That Highly Expresses Factors Driving Pathogenesis of SARS-CoV-2. Front. Cell Dev. Biol. 2020, 8, 783. [Google Scholar] [CrossRef]
- Fahmi, A.; Brügger, M.; Démoulins, T.; Zumkehr, B.; Oliveira Esteves, B.I.; Bracher, L.; Wotzkow, C.; Blank, F.; Thiel, V.; Baud, D.; et al. SARS-CoV-2 Can Infect and Propagate in Human Placenta Explants. Cell Rep. Med. 2021, 2, 100456. [Google Scholar] [CrossRef]
- Lu-Culligan, A.; Chavan, A.R.; Vijayakumar, P.; Irshaid, L.; Courchaine, E.M.; Milano, K.M.; Tang, Z.; Pope, S.D.; Song, E.; Vogels, C.B.F.; et al. Maternal Respiratory SARS-CoV-2 Infection in Pregnancy Is Associated with a Robust Inflammatory Response at the Maternal-Fetal Interface. Med 2021, 2, 591–610. [Google Scholar] [CrossRef]
- Verma, S.; Joshi, C.S.; Silverstein, R.B.; He, M.; Carter, E.B.; Mysorekar, I.U. SARS-CoV-2 Colonization of Maternal and Fetal Cells of the Human Placenta Promotes Alteration of Local Renin-Angiotensin System. Med 2021, 2, 575–590. [Google Scholar] [CrossRef]
- Allotey, J.; Chatterjee, S.; Kew, T.; Gaetano, A.; Stallings, E.; Fernández-García, S.; Yap, M.; Sheikh, J.; Lawson, H.; Coomar, D.; et al. SARS-CoV-2 Positivity in Offspring and Timing of Mother-to-Child Transmission: Living Systematic Review and Meta-Analysis. BMJ 2022, 376, e067696. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Definition and Categorization of the Timing of Mother-to-Child Transmission of SARS-CoV-2: Scientific Brief, 8 February 2021; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Babaei, R.; Bokharaei-Salim, F.; Khanaliha, K.; Kiani, S.J.; Marjani, A.; Garshasbi, S.; Dehghani-Dehej, F.; Chavoshpour, S. Prevalence of SARS-CoV-2 Infection in Neonates Born to Mothers or Relatives with COVID-19. BMC Infect. Dis. 2022, 22, 730. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Serrano, D.A.; Roy-Vallejo, E.; Zurita Cruz, N.D.; Martín Ramírez, A.; Rodríguez-García, S.C.; Arevalillo-Fernández, N.; Galván-Román, J.M.; Fontán García-Rodrigo, L.; Vega-Piris, L.; Chicot Llano, M.; et al. Detection of SARS-CoV-2 RNA in Serum Is Associated with Increased Mortality Risk in Hospitalized COVID-19 Patients. Sci. Rep. 2021, 11, 13134. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Flores, V.; Romero, R.; Xu, Y.; Theis, K.R.; Arenas-Hernandez, M.; Miller, D.; Peyvandipour, A.; Bhatti, G.; Galaz, J.; Gershater, M.; et al. Maternal-Fetal Immune Responses in Pregnant Women Infected with SARS-CoV-2. Nat. Commun. 2022, 13, 320. [Google Scholar] [CrossRef] [PubMed]
- Suhren, J.-T.; Meinardus, A.; Hussein, K.; Schaumann, N. Meta-Analysis on COVID-19-Pregnancy-Related Placental Pathologies Shows No Specific Pattern. Placenta 2022, 117, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Dang, D.; Wang, L.; Zhang, C.; Li, Z.; Wu, H. Potential Effects of SARS-CoV-2 Infection during Pregnancy on Fetuses and Newborns Are Worthy of Attention. J. Obstet. Gynaecol. Res. 2020, 46, 1951–1957. [Google Scholar] [CrossRef]
- Foo, S.-S.; Cambou, M.C.; Mok, T.; Fajardo, V.M.; Jung, K.L.; Fuller, T.; Chen, W.; Kerin, T.; Mei, J.; Bhattacharya, D.; et al. The Systemic Inflammatory Landscape of COVID-19 in Pregnancy: Extensive Serum Proteomic Profiling of Mother-Infant Dyads with in Utero SARS-CoV-2. Cell Rep. Med. 2021, 2, 100453. [Google Scholar] [CrossRef]
- Ge, E.; Li, Y.; Wu, S.; Candido, E.; Wei, X. Association of Pre-Existing Comorbidities with Mortality and Disease Severity among 167,500 Individuals with COVID-19 in Canada: A Population-Based Cohort Study. PLoS ONE 2021, 16, e0258154. [Google Scholar] [CrossRef]
- Gutierrez, J.P.; Bertozzi, S.M. Non-Communicable Diseases and Inequalities Increase Risk of Death among COVID-19 Patients in Mexico. PLoS ONE 2020, 15, e0240394. [Google Scholar] [CrossRef]
- Pardhan, S.; Wood, S.; Vaughan, M.; Trott, M. The Risk of COVID-19 Related Hospitalsation, Intensive Care Unit Admission and Mortality in People with Underlying Asthma or COPD: A Systematic Review and Meta-Analysis. Front. Med. 2021, 8, 668808. [Google Scholar] [CrossRef]
- Mahamat-Saleh, Y.; Fiolet, T.; Rebeaud, M.E.; Mulot, M.; Guihur, A.; Fatouhi, D.E.; Laouali, N.; Peiffer-Smadja, N.; Aune, D.; Severi, G. Diabetes, Hypertension, Body Mass Index, Smoking and COVID-19-Related Mortality: A Systematic Review and Meta-Analysis of Observational Studies. BMJ Open 2021, 11, e052777. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Kim, S.R.; Kim, M.-N.; Shim, W.J.; Park, S.-M. Impact of Cardiovascular Disease and Risk Factors on Fatal Outcomes in Patients with COVID-19 According to Age: A Systematic Review and Meta-Analysis. Heart 2021, 107, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Zhang, J.; Zhu, Y.; Liu, L.; Liu, Y.; He, Q. Mortality in Chronic Kidney Disease Patients with COVID-19: A Systematic Review and Meta-Analysis. Int. Urol. Nephrol. 2021, 53, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Yang, Y.; Zhang, J. Obesity Is Associated with Severe Disease and Mortality in Patients with Coronavirus Disease 2019 (COVID-19): A Meta-Analysis. BMC Public Health 2021, 21, 1505. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, M.-R.; Amin, R.; Maher, A.; Bahadorimonfared, A.; Janbazi, S.; Hannani, K.; Kolahi, A.-A.; Zali, A.-R. Sociodemographic Determinants and Clinical Risk Factors Associated with COVID-19 Severity: A Cross-Sectional Analysis of over 200,000 Patients in Tehran, Iran. BMC Infect. Dis. 2021, 21, 474. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhuang, Q.; Chiang, J.; Tan, S.H.; Chua, G.W.Y.; Xie, C.; Chua, M.L.K.; Soon, Y.Y.; Yang, V.S. Impact of Cancer Diagnoses on the Outcomes of Patients with COVID-19: A Systematic Review and Meta-Analysis. BMJ Open 2022, 12, e044661. [Google Scholar] [CrossRef]
- The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 14 June 2022).
- Gerayeli, F.V.; Milne, S.; Cheung, C.; Li, X.; Yang, C.W.T.; Tam, A.; Choi, L.H.; Bae, A.; Sin, D.D. COPD and the Risk of Poor Outcomes in COVID-19: A Systematic Review and Meta-Analysis. EClinicalMedicine 2021, 33, 100789. [Google Scholar] [CrossRef]
- Agustí, A.; Hogg, J.C. Update on the Pathogenesis of Chronic Obstructive Pulmonary Disease. N. Engl. J. Med. 2019, 381, 1248–1256. [Google Scholar] [CrossRef]
- Higham, A.; Mathioudakis, A.; Vestbo, J.; Singh, D. COVID-19 and COPD: A Narrative Review of the Basic Science and Clinical Outcomes. Eur. Respir. Rev. 2020, 29, 200199. [Google Scholar] [CrossRef]
- Mallia, P.; Message, S.D.; Gielen, V.; Contoli, M.; Gray, K.; Kebadze, T.; Aniscenko, J.; Laza-Stanca, V.; Edwards, M.R.; Slater, L.; et al. Experimental Rhinovirus Infection as a Human Model of Chronic Obstructive Pulmonary Disease Exacerbation. Am. J. Respir. Crit. Care Med. 2011, 183, 734–742. [Google Scholar] [CrossRef] [Green Version]
- Barnes, P.J. Inflammatory Mechanisms in Patients with Chronic Obstructive Pulmonary Disease. J. Allergy Clin. Immunol. 2016, 138, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zeng, M.; Wang, H.; Qin, C.; Hou, H.; Sun, Z.; Xu, S.; Wang, G.; Guo, C.; Deng, Y.; et al. Distinct Effects of Asthma and COPD Comorbidity on Disease Expression and Outcome in Patients with COVID-19. Allergy 2021, 76, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.M.; Yang, C.X.; Tam, A.; Shaipanich, T.; Hackett, T.-L.; Singhera, G.K.; Dorscheid, D.R.; Sin, D.D. ACE-2 Expression in the Small Airway Epithelia of Smokers and COPD Patients: Implications for COVID-19. Eur. Respir. J. 2020, 55, 2000688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milne, S.; Yang, C.X.; Timens, W.; Bossé, Y.; Sin, D.D. SARS-CoV-2 Receptor ACE2 Gene Expression and RAAS Inhibitors. Lancet Respir. Med. 2020, 8, e50–e51. [Google Scholar] [CrossRef] [PubMed]
- Mulpuru, S.; Li, L.; Ye, L.; Hatchette, T.; Andrew, M.K.; Ambrose, A.; Boivin, G.; Bowie, W.; Chit, A.; Santos, G.D.; et al. Effectiveness of Influenza Vaccination on Hospitalizations and Risk Factors for Severe Outcomes in Hospitalized Patients with COPD. CHEST 2019, 155, 69–78. [Google Scholar] [CrossRef]
- Restrepo, M.I.; Mortensen, E.M.; Pugh, J.A.; Anzueto, A. COPD Is Associated with Increased Mortality in Patients with Community-Acquired Pneumonia. Eur. Respir. J. 2006, 28, 346–351. [Google Scholar] [CrossRef] [Green Version]
- Bafadhel, M.; McKenna, S.; Terry, S.; Mistry, V.; Reid, C.; Haldar, P.; McCormick, M.; Haldar, K.; Kebadze, T.; Duvoix, A.; et al. Acute Exacerbations of Chronic Obstructive Pulmonary Disease: Identification of Biologic Clusters and Their Biomarkers. Am. J. Respir. Crit. Care Med. 2011, 184, 662–671. [Google Scholar] [CrossRef]
- George, S.N.; Garcha, D.S.; Mackay, A.J.; Patel, A.R.C.; Singh, R.; Sapsford, R.J.; Donaldson, G.C.; Wedzicha, J.A. Human Rhinovirus Infection during Naturally Occurring COPD Exacerbations. Eur. Respir. J. 2014, 44, 87–96. [Google Scholar] [CrossRef]
- Wilkinson, T.M.A.; Hurst, J.R.; Perera, W.R.; Wilks, M.; Donaldson, G.C.; Wedzicha, J.A. Effect of Interactions between Lower Airway Bacterial and Rhinoviral Infection in Exacerbations of COPD. Chest 2006, 129, 317–324. [Google Scholar] [CrossRef]
- Mathioudakis, A.G.; Janssens, W.; Sivapalan, P.; Singanayagam, A.; Dransfield, M.T.; Jensen, J.-U.S.; Vestbo, J. Acute Exacerbations of Chronic Obstructive Pulmonary Disease: In Search of Diagnostic Biomarkers and Treatable Traits. Thorax 2020, 75, 520–527. [Google Scholar] [CrossRef] [Green Version]
- Vanfleteren, L.E.G.W.; Spruit, M.A.; Wouters, E.F.M.; Franssen, F.M.E. Management of Chronic Obstructive Pulmonary Disease beyond the Lungs. Lancet Respir. Med. 2016, 4, 911–924. [Google Scholar] [CrossRef]
- Terzano, C.; Colamesta, V.; Unim, B.; Romani, S.; Meneghini, A.; Volpe, G.; La Torre, G. Chronic Obstructive Pulmonary Disease (COPD) Exacerbation: Impact of Comorbidities on Length and Costs during Hospitalization. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3680–3689. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.E.; McAvay, G.J.; Allore, H.G.; Stamm, J.A.; Simonelli, P.F. Contributions of COPD, Asthma, and Ten Comorbid Conditions to Health Care Utilization and Patient-Centered Outcomes among US Adults with Obstructive Airway Disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2017, 12, 2515–2522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonso, A.S.M.; Verhamme, K.M.C.; Sturkenboom, M.C.J.M.; Brusselle, G.G.O. COPD in the General Population: Prevalence, Incidence and Survival. Respir. Med. 2011, 105, 1872–1884. [Google Scholar] [CrossRef] [PubMed]
- Podolanczuk, A.J.; Richeldi, L. COVID-19 and Interstitial Lung Disease: Keep Them Separate. Am. J. Respir. Crit. Care Med. 2020, 202, 1614–1616. [Google Scholar] [CrossRef]
- Castelino, F.V.; Varga, J. Interstitial Lung Disease in Connective Tissue Diseases: Evolving Concepts of Pathogenesis and Management. Arthritis Res. Ther. 2010, 12, 213. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Choi, H.; Yang, B.; Lee, S.-K.; Park, T.S.; Park, D.W.; Moon, J.-Y.; Kim, T.-H.; Sohn, J.W.; Yoon, H.J.; et al. Interstitial Lung Disease Increases Susceptibility to and Severity of COVID-19. Eur. Respir. J. 2021, 58, 2004125. [Google Scholar] [CrossRef]
- Esposito, A.J.; Menon, A.A.; Ghosh, A.J.; Putman, R.K.; Fredenburgh, L.E.; El-Chemaly, S.Y.; Goldberg, H.J.; Baron, R.M.; Hunninghake, G.M.; Doyle, T.J. Increased Odds of Death for Patients with Interstitial Lung Disease and COVID-19: A Case–Control Study. Am. J. Respir. Crit. Care Med. 2020, 202, 1710–1713. [Google Scholar] [CrossRef]
- Ouyang, L.; Gong, J.; Yu, M. Pre-Existing Interstitial Lung Disease in Patients with Coronavirus Disease 2019: A Meta-Analysis. Int. Immunopharmacol. 2021, 100, 108145. [Google Scholar] [CrossRef]
- Drake, T.M.; Docherty, A.B.; Harrison, E.M.; Quint, J.K.; Adamali, H.; Agnew, S.; Babu, S.; Barber, C.M.; Barratt, S.; Bendstrup, E.; et al. Outcome of Hospitalization for COVID-19 in Patients with Interstitial Lung Disease. An International Multicenter Study. Am. J. Respir. Crit. Care Med. 2020, 202, 1656–1665. [Google Scholar] [CrossRef]
- Lee, S.C.; Son, K.J.; Han, C.H.; Jung, J.Y.; Park, S.C. Impact of Comorbid Asthma on Severity of Coronavirus Disease (COVID-19). Sci. Rep. 2020, 10, 21805. [Google Scholar] [CrossRef] [PubMed]
- Wark, P.A.B.; Pathinayake, P.S.; Kaiko, G.; Nichol, K.; Ali, A.; Chen, L.; Sutanto, E.N.; Garratt, L.W.; Sohal, S.S.; Lu, W.; et al. ACE2 Expression Is Elevated in Airway Epithelial Cells from Older and Male Healthy Individuals but Reduced in Asthma. Respirol. Carlton Vic 2021, 26, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.J.; Busse, W.W.; Bacharier, L.B.; Kattan, M.; O’Connor, G.T.; Wood, R.A.; Visness, C.M.; Durham, S.R.; Larson, D.; Esnault, S.; et al. Association of Respiratory Allergy, Asthma, and Expression of the SARS-CoV-2 Receptor ACE2. J. Allergy Clin. Immunol. 2020, 146, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Skevaki, C.; Karsonova, A.; Karaulov, A.; Fomina, D.; Xie, M.; Chinthrajah, S.; Nadeau, K.C.; Renz, H. SARS-CoV-2 Infection and COVID-19 in Asthmatics: A Complex Relationship. Nat. Rev. Immunol. 2021, 21, 202–203. [Google Scholar] [CrossRef]
- Ho, K.S.; Howell, D.; Rogers, L.; Narasimhan, B.; Verma, H.; Steiger, D. The Relationship between Asthma, Eosinophilia, and Outcomes in Coronavirus Disease 2019 Infection. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2021, 127, 42–48. [Google Scholar] [CrossRef]
- Peltola, V.; Jartti, T.; Putto-Laurila, A.; Mertsola, J.; Vainionpää, R.; Waris, M.; Hyypiä, T.; Ruuskanen, O. Rhinovirus Infections in Children: A Retrospective and Prospective Hospital-Based Study. J. Med. Virol. 2009, 81, 1831–1838. [Google Scholar] [CrossRef]
- Nicholson, K.G.; Kent, J.; Ireland, D.C. Respiratory Viruses and Exacerbations of Asthma in Adults. Br. Med. J. 1993, 307, 982–986. [Google Scholar] [CrossRef] [Green Version]
- Johnston, S.L.; Pattemore, P.K.; Sanderson, G.; Smith, S.; Lampe, F.; Josephs, L.; Symington, P.; Toole, S.O.; Myint, S.H.; Tyrrell, D.A.J.; et al. Community Study of Role of Viral Infections in Exacerbations of Asthma in 9-11 Year Old Children. BMJ 1995, 310, 1225–1229. [Google Scholar] [CrossRef] [Green Version]
- Contoli, M.; Message, S.D.; Laza-Stanca, V.; Edwards, M.R.; Wark, P.A.B.; Bartlett, N.W.; Kebadze, T.; Mallia, P.; Stanciu, L.A.; Parker, H.L.; et al. Role of Deficient Type III Interferon-λ Production in Asthma Exacerbations. Nat. Med. 2006, 12, 1023–1026. [Google Scholar] [CrossRef]
- Gill, M.A.; Bajwa, G.; George, T.A.; Dong, C.C.; Dougherty, I.I.; Jiang, N.; Gan, V.N.; Gruchalla, R.S. Counterregulation between the FcεRI Pathway and Antiviral Responses in Human Plasmacytoid Dendritic Cells. J. Immunol. 2010, 184, 5999–6006. [Google Scholar] [CrossRef] [Green Version]
- Gonzales-van Horn, S.R.; David Farrar, J. Interferon at the Crossroads of Allergy and Viral Infections. J. Leukoc. Biol. 2015, 98, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Coleman, B.L.; Fadel, S.A.; Fitzpatrick, T.; Thomas, S.-M. Risk Factors for Serious Outcomes Associated with Influenza Illness in High- versus Low- and Middle-Income Countries: Systematic Literature Review and Meta-Analysis. Influenza Other Respir. Viruses 2018, 12, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.M.; Koh, H.Y.; Moon, S.Y.; Yoo, I.K.; Ha, E.K.; You, S.; Kim, S.Y.; Yon, D.K.; Lee, S.W. Allergic Disorders and Susceptibility to and Severity of COVID-19: A Nationwide Cohort Study. J. Allergy Clin. Immunol. 2020, 146, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Hasegawa, K.; Ma, B.; Fujiogi, M.; Camargo, C.A.; Liang, L. Association of Asthma and Its Genetic Predisposition with the Risk of Severe COVID-19. J. Allergy Clin. Immunol. 2020, 146, 327–329.e4. [Google Scholar] [CrossRef] [PubMed]
- World Obesity Atlas 2022. Available online: https://www.worldobesity.org/resources/resource-library/world-obesity-atlas-2022 (accessed on 22 October 2022).
- Petrie, J.R.; Guzik, T.J.; Touyz, R.M. Diabetes, Hypertension, and Cardiovascular Disease: Clinical Insights and Vascular Mechanisms. Can. J. Cardiol. 2018, 34, 575–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madjid, M.; Safavi-Naeini, P.; Solomon, S.D.; Vardeny, O. Potential Effects of Coronaviruses on the Cardiovascular System: A Review. JAMA Cardiol. 2020, 5, 831–840. [Google Scholar] [CrossRef] [Green Version]
- Badawi, A.; Ryoo, S.G. Prevalence of Comorbidities in the Middle East Respiratory Syndrome Coronavirus (MERS-CoV): A Systematic Review and Meta-Analysis. Int. J. Infect. Dis. 2016, 49, 129–133. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.K.; Feng, Y.; Yuan, M.Y.; Yuan, S.Y.; Fu, H.J.; Wu, B.Y.; Sun, G.Z.; Yang, G.R.; Zhang, X.L.; Wang, L.; et al. Plasma Glucose Levels and Diabetes Are Independent Predictors for Mortality and Morbidity in Patients with SARS. Diabet. Med. J. Br. Diabet. Assoc. 2006, 23, 623–628. [Google Scholar] [CrossRef]
- Kesavadev, J.; Misra, A.; Saboo, B.; Aravind, S.R.; Hussain, A.; Czupryniak, L.; Raz, I. Blood Glucose Levels Should Be Considered as a New Vital Sign Indicative of Prognosis during Hospitalization. Diabetes Metab. Syndr. 2021, 15, 221–227. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, S.; Zhang, L.; Dong, Y.; Zhang, S. Admission Fasting Blood Glucose Predicts 30-Day Poor Outcome in Patients Hospitalized for COVID-19 Pneumonia. Diabetes Obes. Metab. 2020, 22, 1955–1957. [Google Scholar] [CrossRef]
- Wang, S.; Ma, P.; Zhang, S.; Song, S.; Wang, Z.; Ma, Y.; Xu, J.; Wu, F.; Duan, L.; Yin, Z.; et al. Fasting Blood Glucose at Admission Is an Independent Predictor for 28-Day Mortality in Patients with COVID-19 without Previous Diagnosis of Diabetes: A Multi-Centre Retrospective Study. Diabetologia 2020, 63, 2102–2111. [Google Scholar] [CrossRef] [PubMed]
- Böhm, M.; Frey, N.; Giannitsis, E.; Sliwa, K.; Zeiher, A.M. Coronavirus Disease 2019 (COVID-19) and Its Implications for Cardiovascular Care: Expert Document from the German Cardiac Society and the World Heart Federation. Clin. Res. Cardiol. 2020, 109, 1446–1459. [Google Scholar] [CrossRef] [PubMed]
- Kai, H.; Kai, M. Interactions of Coronaviruses with ACE2, Angiotensin II, and RAS Inhibitors—Lessons from Available Evidence and Insights into COVID-19. Hypertens. Res. 2020, 43, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Zhao, Q.; Kumar, R.; Bai, C.; Deng, Y.; Wan, B. Impact of Cardiovascular and Metabolic Diseases on the Severity of COVID-19: A Systematic Review and Meta-Analysis. Aging 2020, 12, 23409–23421. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kompoti, M. Obesity and Infection. Lancet Infect. Dis. 2006, 6, 438–446. [Google Scholar] [CrossRef]
- Poulain, M.; Doucet, M.; Major, G.C.; Drapeau, V.; Sériès, F.; Boulet, L.-P.; Tremblay, A.; Maltais, F. The Effect of Obesity on Chronic Respiratory Diseases: Pathophysiology and Therapeutic Strategies. CMAJ 2006, 174, 1293–1299. [Google Scholar] [CrossRef] [Green Version]
- Parameswaran, K.; Todd, D.C.; Soth, M. Altered Respiratory Physiology in Obesity. Can. Respir. J. 2006, 13, 203–210. [Google Scholar] [CrossRef]
- He, H.; Wang, B.; Zhou, M.; Cao, L.; Qiu, W.; Mu, G.; Chen, A.; Yang, S.; Chen, W. Systemic Inflammation Mediates the Associations Between Abdominal Obesity Indices and Lung Function Decline in a Chinese General Population. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Stefan, N. SARS-CoV-2 Fires up Inflammation in Adipose Tissue. Nat. Rev. Endocrinol. 2022, 19, 8–9. [Google Scholar] [CrossRef]
- Sibbel, S.; Sato, R.; Hunt, A.; Turenne, W.; Brunelli, S.M. The Clinical and Economic Burden of Pneumonia in Patients Enrolled in Medicare Receiving Dialysis: A Retrospective, Observational Cohort Study. BMC Nephrol. 2016, 17, 199. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Galdamez, D.R.; González-Block, M.Á.; Romo-Dueñas, D.K.; Lima-Morales, R.; Hernández-Vicente, I.A.; Lumbreras-Guzmán, M.; Méndez-Hernández, P. Increased Risk of Hospitalization and Death in Patients with COVID-19 and Pre-Existing Noncommunicable Diseases and Modifiable Risk Factors in Mexico. Arch. Med. Res. 2020, 51, 683–689. [Google Scholar] [CrossRef]
- Sun, J.; Zhu, A.; Li, H.; Zheng, K.; Zhuang, Z.; Chen, Z.; Shi, Y.; Zhang, Z.; Chen, S.-B.; Liu, X.; et al. Isolation of Infectious SARS-CoV-2 from Urine of a COVID-19 Patient. Emerg. Microbes Infect. 2020, 9, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Legrand, M.; Bell, S.; Forni, L.; Joannidis, M.; Koyner, J.L.; Liu, K.; Cantaluppi, V. Pathophysiology of COVID-19-Associated Acute Kidney Injury. Nat. Rev. Nephrol. 2021, 17, 751–764. [Google Scholar] [CrossRef]
- Sandoval, M.; Nguyen, D.T.; Vahidy, F.S.; Graviss, E.A. Risk Factors for Severity of COVID-19 in Hospital Patients Age 18–29 Years. PLoS ONE 2021, 16, e0255544. [Google Scholar] [CrossRef] [PubMed]
- Sunjaya, A.P.; Allida, S.M.; Di Tanna, G.L.; Jenkins, C. Asthma and Risk of Infection, Hospitalization, ICU Admission and Mortality from COVID-19: Systematic Review and Meta-Analysis. J. Asthma 2022, 59, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Xu, J.; Hou, H.; Yang, H.; Wang, Y. Impact of Asthma on COVID-19 Mortality in the United States: Evidence Based on a Meta-Analysis. Int. Immunopharmacol. 2022, 102, 108390. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Xu, J.; Li, Y.; Wang, Y.; Yang, H. The Association of Asthma with COVID-19 Mortality: An Updated Meta-Analysis Based on Adjusted Effect Estimates. J. Allergy Clin. Immunol. Pract. 2021, 9, 3944–3968. [Google Scholar] [CrossRef]
- Hussein, M.H.; Elshazli, R.M.; Attia, A.S.; Nguyen, T.P.; Aboueisha, M.; Munshi, R.; Toraih, E.A.; Fawzy, M.S.; Kandil, E. Asthma and COVID-19; Different Entities, Same Outcome: A Meta-Analysis of 107,983 Patients. J. Asthma 2022, 59, 851–858. [Google Scholar] [CrossRef]
- Liu, S.; Cao, Y.; Du, T.; Zhi, Y. Prevalence of Comorbid Asthma and Related Outcomes in COVID-19: A Systematic Review and Meta-Analysis. J. Allergy Clin. Immunol. Pract. 2021, 9, 693–701. [Google Scholar] [CrossRef]
- Shi, L.; Xu, J.; Xiao, W.; Wang, Y.; Jin, Y.; Chen, S.; Duan, G.; Yang, H.; Wang, Y. Asthma in Patients with Coronavirus Disease 2019: A Systematic Review and Meta-Analysis. Ann. Allergy. Asthma. Immunol. 2021, 126, 524–534. [Google Scholar] [CrossRef]
- Wu, T.; Yu, P.; Li, Y.; Wang, J.; Li, Z.; Qiu, J.; Cui, L.; Mou, Y.; Sun, Y. Asthma Does Not Influence the Severity of COVID-19: A Meta-Analysis. J. Asthma 2022, 59, 1188–1194. [Google Scholar] [CrossRef] [PubMed]
- Otunla, A.; Rees, K.; Dennison, P.; Hobbs, R.; Suklan, J.; Schofield, E.; Gunnell, J.; Mighiu, A.; Hartmann-Boyce, J. Risks of Infection, Hospital and ICU Admission, and Death from COVID-19 in People with Asthma: Systematic Review and Meta-Analyses. BMJ Evid.-Based Med. 2022, 27, 263–273. [Google Scholar] [CrossRef]
- Sunjaya, A.P.; Allida, S.M.; Tanna, G.L.D.; Jenkins, C.R. Asthma and COVID-19 Risk: A Systematic Review and Meta-Analysis. Eur. Respir. J. 2022, 59, 2101209. [Google Scholar] [CrossRef] [PubMed]
- Reyes, F.M.; Hache-Marliere, M.; Karamanis, D.; Berto, C.G.; Estrada, R.; Langston, M.; Ntaios, G.; Gulani, P.; Shah, C.D.; Palaiodimos, L. Assessment of the Association of COPD and Asthma with In-Hospital Mortality in Patients with COVID-19. A Systematic Review, Meta-Analysis, and Meta-Regression Analysis. J. Clin. Med. 2021, 10, 2087. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Bacon, S.; Evans, S.J.; Bates, C.J.; Rentsch, C.T.; MacKenna, B.; Tomlinson, L.; Walker, A.J.; Schultze, A.; Morton, C.E.; et al. Factors Associated with Deaths Due to COVID-19 versus Other Causes: Population-Based Cohort Analysis of UK Primary Care Data and Linked National Death Registrations within the OpenSAFELY Platform. Lancet Reg. Health—Eur. 2021, 6, 100109. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors Associated with COVID-19-Related Death Using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Szarpak, L.; Mierzejewska, M.; Jurek, J.; Kochanowska, A.; Gasecka, A.; Truszewski, Z.; Pruc, M.; Blek, N.; Rafique, Z.; Filipiak, K.J.; et al. Effect of Coronary Artery Disease on COVID-19—Prognosis and Risk Assessment: A Systematic Review and Meta-Analysis. Biology 2022, 11, 221. [Google Scholar] [CrossRef] [PubMed]
- Hessami, A.; Shamshirian, A.; Heydari, K.; Pourali, F.; Alizadeh-Navaei, R.; Moosazadeh, M.; Abrotan, S.; Shojaie, L.; Sedighi, S.; Shamshirian, D.; et al. Cardiovascular Diseases Burden in COVID-19: Systematic Review and Meta-Analysis. Am. J. Emerg. Med. 2021, 46, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, J.; Ma, F.-Z.; Li, J.; Xue, S.; Xu, Z.-G. The Common Risk Factors for Progression and Mortality in COVID-19 Patients: A Meta-Analysis. Arch. Virol. 2021, 166, 2071–2087. [Google Scholar] [CrossRef]
- Loffi, M.; Piccolo, R.; Regazzoni, V.; Tano, G.D.; Moschini, L.; Robba, D.; Quinzani, F.; Esposito, G.; Franzone, A.; Danzi, G.B. Coronary Artery Disease in Patients Hospitalised with Coronavirus Disease 2019 (COVID-19) Infection. Open Heart 2020, 7, e001428. [Google Scholar] [CrossRef]
- Atkins, J.L.; Masoli, J.A.H.; Delgado, J.; Pilling, L.C.; Kuo, C.-L.; Kuchel, G.A.; Melzer, D. Preexisting Comorbidities Predicting COVID-19 and Mortality in the UK Biobank Community Cohort. J. Gerontol. Ser. A 2020, 75, 2224–2230. [Google Scholar] [CrossRef]
- Singh, S.; Khan, A. Clinical Characteristics and Outcomes of Coronavirus Disease 2019 among Patients with Preexisting Liver Disease in the United States: A Multicenter Research Network Study. Gastroenterology 2020, 159, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Hassan, L.; Peek, N.; Lovell, K.; Carvalho, A.F.; Solmi, M.; Stubbs, B.; Firth, J. Disparities in COVID-19 Infection, Hospitalisation and Death in People with Schizophrenia, Bipolar Disorder, and Major Depressive Disorder: A Cohort Study of the UK Biobank. Mol. Psychiatry 2022, 27, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Galiero, R.; Pafundi, P.C.; Simeon, V.; Rinaldi, L.; Perrella, A.; Vetrano, E.; Caturano, A.; Alfano, M.; Beccia, D.; Nevola, R.; et al. Impact of Chronic Liver Disease upon Admission on COVID-19 in-Hospital Mortality: Findings from COVOCA Study. PLoS ONE 2020, 15, e0243700. [Google Scholar] [CrossRef] [PubMed]
- Gimeno-Miguel, A.; Bliek-Bueno, K.; Poblador-Plou, B.; Carmona-Pírez, J.; Poncel-Falcó, A.; González-Rubio, F.; Ioakeim-Skoufa, I.; Pico-Soler, V.; Aza-Pascual-Salcedo, M.; Prados-Torres, A.; et al. Chronic Diseases Associated with Increased Likelihood of Hospitalization and Mortality in 68,913 COVID-19 Confirmed Cases in Spain: A Population-Based Cohort Study. PLoS ONE 2021, 16, e0259822. [Google Scholar] [CrossRef]
- Hashemi, N.; Viveiros, K.; Redd, W.D.; Zhou, J.C.; McCarty, T.R.; Bazarbashi, A.N.; Hathorn, K.E.; Wong, D.; Njie, C.; Shen, L.; et al. Impact of Chronic Liver Disease on Outcomes of Hospitalized Patients with COVID-19: A Multicentre United States Experience. Liver Int. 2020, 40, 2515–2521. [Google Scholar] [CrossRef]
- Khan, M.M.A.; Khan, M.N.; Mustagir, M.G.; Rana, J.; Islam, M.S.; Kabir, M.I. Effects of Underlying Morbidities on the Occurrence of Deaths in COVID-19 Patients: A Systematic Review and Meta-Analysis. J. Glob. Health 2020, 10, 020503. [Google Scholar] [CrossRef]
- Yin, T.; Li, Y.; Ying, Y.; Luo, Z. Prevalence of Comorbidity in Chinese Patients with COVID-19: Systematic Review and Meta-Analysis of Risk Factors. BMC Infect. Dis. 2021, 21, 200. [Google Scholar] [CrossRef]
- Wang, X.; Fang, X.; Cai, Z.; Wu, X.; Gao, X.; Min, J.; Wang, F. Comorbid Chronic Diseases and Acute Organ Injuries Are Strongly Correlated with Disease Severity and Mortality among COVID-19 Patients: A Systemic Review and Meta-Analysis. Research 2020, 2020, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, A.V.; Kumar, P.; Tevethia, H.V.; Premkumar, M.; Arab, J.P.; Candia, R.; Talukdar, R.; Sharma, M.; Qi, X.; Rao, P.N.; et al. Systematic Review with Meta-Analysis: Liver Manifestations and Outcomes in COVID-19. Aliment. Pharmacol. Ther. 2020, 52, 584–599. [Google Scholar] [CrossRef]
- Lee, Y.R.; Kang, M.K.; Song, J.E.; Kim, H.J.; Kweon, Y.O.; Tak, W.Y.; Jang, S.Y.; Park, J.G.; Lee, C.; Hwang, J.S.; et al. Clinical Outcomes of Coronavirus Disease 2019 in Patients with Pre-Existing Liver Diseases: A Multicenter Study in South Korea. Clin. Mol. Hepatol. 2020, 26, 562–576. [Google Scholar] [CrossRef]
- Treskova-Schwarzbach, M.; Haas, L.; Reda, S.; Pilic, A.; Borodova, A.; Karimi, K.; Koch, J.; Nygren, T.; Scholz, S.; Schönfeld, V.; et al. Pre-Existing Health Conditions and Severe COVID-19 Outcomes: An Umbrella Review Approach and Meta-Analysis of Global Evidence. BMC Med. 2021, 19, 212. [Google Scholar] [CrossRef]
- Frager, S.Z.; Szymanski, J.; Schwartz, J.M.; Massoumi, H.S.; Kinkhabwala, M.; Wolkoff, A.W. Hepatic Predictors of Mortality in Severe Acute Respiratory Syndrome Coronavirus 2: Role of Initial Aspartate Aminotransferase/Alanine Aminotransferase and Preexisting Cirrhosis. Hepatol. Commun. 2021, 5, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Adeniji, N.; Latt, N.; Kumar, S.; Bloom, P.P.; Aby, E.S.; Perumalswami, P.; Roytman, M.; Li, M.; Vogel, A.S.; et al. Predictors of Outcomes of COVID-19 in Patients with Chronic Liver Disease: US Multi-Center Study. Clin. Gastroenterol. Hepatol. 2021, 19, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Jeon, D.; Son, M.; Choi, J. Impact of Liver Cirrhosis on the Clinical Outcomes of Patients with COVID-19: A Nationwide Cohort Study of Korea. Korean J. Intern. Med. 2021, 36, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Mahamid, M.; Nseir, W.; Khoury, T.; Mahamid, B.; Nubania, A.; Sub-Laban, K.; Schifter, J.; Mari, A.; Sbeit, W.; Goldin, E. Nonalcoholic Fatty Liver Disease Is Associated with COVID-19 Severity Independently of Metabolic Syndrome: A Retrospective Case-Control Study. Eur. J. Gastroenterol. Hepatol. 2021, 33, 1578–1581. [Google Scholar] [CrossRef]
- Gao, F.; Zheng, K.I.; Wang, X.; Yan, H.; Sun, Q.; Pan, K.; Wang, T.; Chen, Y.; George, J.; Zheng, M. Metabolic Associated Fatty Liver Disease Increases Coronavirus Disease 2019 Disease Severity in Nondiabetic Patients. J. Gastroenterol. Hepatol. 2021, 36, 204–207. [Google Scholar] [CrossRef]
- Ji, D.; Qin, E.; Xu, J.; Zhang, D.; Cheng, G.; Wang, Y.; Lau, G. Non-Alcoholic Fatty Liver Diseases in Patients with COVID-19: A Retrospective Study. J. Hepatol. 2020, 73, 451–453. [Google Scholar] [CrossRef]
- Pan, L.; Huang, P.; Xie, X.; Xu, J.; Guo, D.; Jiang, Y. Metabolic Associated Fatty Liver Disease Increases the Severity of COVID-19: A Meta-Analysis. Dig. Liver Dis. 2021, 53, 153–157. [Google Scholar] [CrossRef]
- Zhou, Y.-J.; Zheng, K.I.; Wang, X.-B.; Sun, Q.-F.; Pan, K.-H.; Wang, T.-Y.; Ma, H.-L.; Chen, Y.-P.; George, J.; Zheng, M.-H. Metabolic-Associated Fatty Liver Disease Is Associated with Severity of COVID-19. Liver Int. 2020, 40, 2160–2163. [Google Scholar] [CrossRef]
- Tao, Z.; Li, Y.; Cheng, B.; Zhou, T.; Gao, Y. Risk of Severe COVID-19 Increased by Metabolic Dysfunction-Associated Fatty Liver Disease. J. Clin. Gastroenterol. 2021, 55, 830–835. [Google Scholar] [CrossRef]
- Hegyi, P.J.; Váncsa, S.; Ocskay, K.; Dembrovszky, F.; Kiss, S.; Farkas, N.; Erőss, B.; Szakács, Z.; Hegyi, P.; Pár, G. Metabolic Associated Fatty Liver Disease Is Associated with an Increased Risk of Severe COVID-19: A Systematic Review with Meta-Analysis. Front. Med. 2021, 8, 626425. [Google Scholar] [CrossRef]
- Singh, A.; Hussain, S.; Antony, B. Non-Alcoholic Fatty Liver Disease and Clinical Outcomes in Patients with COVID-19: A Comprehensive Systematic Review and Meta-Analysis. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-J.; Zheng, K.I.; Wang, X.-B.; Yan, H.-D.; Sun, Q.-F.; Pan, K.-H.; Wang, T.-Y.; Ma, H.-L.; Chen, Y.-P.; George, J.; et al. Younger Patients with MAFLD Are at Increased Risk of Severe COVID-19 Illness: A Multicenter Preliminary Analysis. J. Hepatol. 2020, 73, 719–721. [Google Scholar] [CrossRef] [PubMed]
- Marjot, T.; Moon, A.M.; Cook, J.A.; Abd-Elsalam, S.; Aloman, C.; Armstrong, M.J.; Pose, E.; Brenner, E.J.; Cargill, T.; Catana, M.-A.; et al. Outcomes Following SARS-CoV-2 Infection in Patients with Chronic Liver Disease: An International Registry Study. J. Hepatol. 2021, 74, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Eder, L.; Croxford, R.; Drucker, A.M.; Mendel, A.; Kuriya, B.; Touma, Z.; Johnson, S.R.; Cook, R.; Bernatsky, S.; Haroon, N.; et al. COVID-19 Hospitalizations, Intensive Care Unit Stays, Ventilation, and Death among Patients with Immune-Mediated Inflammatory Diseases Compared to Controls. J. Rheumatol. 2022, 49, 523–530. [Google Scholar] [CrossRef]
- Yang, H.; Xu, J.; Liang, X.; Shi, L.; Wang, Y. Autoimmune Diseases Are Independently Associated with COVID-19 Severity: Evidence Based on Adjusted Effect Estimates. J. Infect. 2021, 82, e23–e26. [Google Scholar] [CrossRef] [PubMed]
- Faye, A.S.; Lee, K.E.; Laszkowska, M.; Kim, J.; Blackett, J.W.; McKenney, A.S.; Krigel, A.; Giles, J.T.; Wang, R.; Bernstein, E.J.; et al. Risk of Adverse Outcomes in Hospitalized Patients with Autoimmune Disease and COVID-19: A Matched Cohort Study from New York City. J. Rheumatol. 2021, 48, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gao, Y.; Zhang, Y.; Shi, S.; Chen, Y.; Tian, J. The Association between Severe or Dead COVID-19 and Autoimmune Diseases: A Systematic Review and Meta-Analysis. J. Infect. 2020, 81, e93–e95. [Google Scholar] [CrossRef]
- Monreal, E.; Sainz de la Maza, S.; Fernández-Velasco, J.I.; Natera-Villalba, E.; Rita, C.G.; Rodríguez-Jorge, F.; Beltrán-Corbellini, Á.; Iturrieta-Zuazo, I.; Rodríguez de Santiago, E.; Espiño, M.; et al. The Impact of Immunosuppression and Autoimmune Disease on Severe Outcomes in Patients Hospitalized with COVID-19. J. Clin. Immunol. 2021, 41, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, J.; Nielsen, J.; Ellingsen, T.; Knudsen, T.; Nielsen, R.; Larsen, M.; Lund, K.; Nørgård, B. Outcome of COVID-19 in Hospitalized Patients with Chronic Inflammatory Diseases. A Population Based National Register Study in Denmark. J. Autoimmun. 2021, 120, 102632. [Google Scholar] [CrossRef]
- Moreno-Torres, V.; de Mendoza, C.; Mellor-Pita, S.; Martínez-Urbistondo, M.; Durán-del Campo, P.; Tutor-Ureta, P.; Vázquez-Comendador, J.-M.; Calderón-Parra, J.; Múñez-Rubio, E.; Ramos-Martínez, A.; et al. Systemic Autoimmune Diseases in Patients Hospitalized with COVID-19 in Spain: A Nation-Wide Registry Study. Viruses 2022, 14, 1631. [Google Scholar] [CrossRef]
- Jung, Y.; Kwon, M.; Choi, H.G. Association between Previous Rheumatoid Arthritis and COVID-19 and Its Severity: A Nationwide Cohort Study in South Korea. BMJ Open 2021, 11, e054753. [Google Scholar] [CrossRef] [PubMed]
- Raiker, R.; DeYoung, C.; Pakhchanian, H.; Ahmed, S.; Kavadichanda, C.; Gupta, L.; Kardeş, S. Outcomes of COVID-19 in Patients with Rheumatoid Arthritis: A Multicenter Research Network Study in the United States. Semin. Arthritis Rheum. 2021, 51, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Nørgård, B.M.; Nielsen, J.; Knudsen, T.; Nielsen, R.G.; Larsen, M.D.; Jølving, L.R.; Kjeldsen, J. Hospitalization for COVID-19 in Patients Treated with Selected Immunosuppressant and Immunomodulating Agents, Compared to the General Population: A Danish Cohort Study. Br. J. Clin. Pharmacol. 2021, 87, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Brodin, R.; Desirée van der Werff, S.; Hedberg, P.; Färnert, A.; Nauclér, P.; Bergman, P.; Requena-Méndez, A. The Association between Pre-Exposure to Glucocorticoids and Other Immunosuppressant Drugs with Severe COVID-19 Outcomes. Clin. Microbiol. Infect. 2022, 28, 1477–1485. [Google Scholar] [CrossRef]
- Rutherford, M.A.; Scott, J.; Karabayas, M.; Antonelou, M.; Gopaluni, S.; Gray, D.; Barrett, J.; Brix, S.R.; Dhaun, N.; McAdoo, S.P.; et al. Risk Factors for Severe Outcomes in Patients with Systemic Vasculitis and COVID-19: A Binational, Registry-Based Cohort Study. Arthritis Rheumatol. 2021, 73, 1713–1719. [Google Scholar] [CrossRef]
- Ahlström, B.; Frithiof, R.; Hultström, M.; Larsson, I.-M.; Strandberg, G.; Lipcsey, M. The Swedish COVID-19 Intensive Care Cohort: Risk Factors of ICU Admission and ICU Mortality. Acta Anaesthesiol. Scand. 2021, 65, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Suárez-García, I.; Perales-Fraile, I.; González-García, A.; Muñoz-Blanco, A.; Manzano, L.; Fabregate, M.; Díez-Manglano, J.; Aizpuru, E.F.; Fernández, F.A.; García, A.G.; et al. In-Hospital Mortality among Immunosuppressed Patients with COVID-19: Analysis from a National Cohort in Spain. PLoS ONE 2021, 16, e0255524. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Ardissino, M.; Reed, T.a.N.; Goodall, J.; Utting, P.; Miscampbell, M.; Condurache, D.G.; Cohen, D.L. Clinical Characteristics and Outcomes of Immunosuppressed Patients Hospitalized with COVID-19: Experience from London. J. Intern. Med. 2021, 289, 385–394. [Google Scholar] [CrossRef]
- Calderón-Parra, J.; Cuervas-Mons, V.; Moreno-Torres, V.; Rubio-Rivas, M.; Blas, P.A.; Pinilla-Llorente, B.; Helguera-Amezua, C.; Jiménez-García, N.; Pesqueira-Fontan, P.-M.; Méndez-Bailón, M.; et al. Influence of Chronic Use of Corticosteroids and Calcineurin Inhibitors on COVID-19 Clinical Outcomes: Analysis of a Nationwide Registry. Int. J. Infect. Dis. 2022, 116, 51–58. [Google Scholar] [CrossRef]
- Pablos, J.L.; Galindo, M.; Carmona, L.; Lledó, A.; Retuerto, M.; Blanco, R.; Gonzalez-Gay, M.A.; Martinez-Lopez, D.; Castrejón, I.; Alvaro-Gracia, J.M.; et al. Clinical Outcomes of Hospitalised Patients with COVID-19 and Chronic Inflammatory and Autoimmune Rheumatic Diseases: A Multicentric Matched Cohort Study. Ann. Rheum. Dis. 2020, 79, 1544–1549. [Google Scholar] [CrossRef]
- El Fakih, R.; Haroon, A.; Alfraih, F.; Al-Khabori, M.K.; Alzahrani, M.; Alhuraiji, A.; Hamadah, A.; AlJohani, N.I.; Alahmari, B.; Essa, M.F.; et al. Clinical Course and Outcomes of COVID-19 in Hematopoietic Cell Transplant Patients, a Regional Report from the Middle East. Bone Marrow Transplant. 2021, 56, 2144–2151. [Google Scholar] [CrossRef]
- Sun, J.; Patel, R.C.; Zheng, Q.; Madhira, V.; Olex, A.L.; Islam, J.Y.; French, E.; Chiang, T.P.-Y.; Akselrod, H.; Moffitt, R.; et al. COVID-19 Disease Severity among People with HIV Infection or Solid Organ Transplant in the United States: A Nationally-Representative, Multicenter, Observational Cohort Study. medRxiv 2021. [Google Scholar] [CrossRef]
- Gatti, M.; Rinaldi, M.; Bussini, L.; Bonazzetti, C.; Pascale, R.; Pasquini, Z.; Faní, F.; Pinho Guedes, M.N.; Azzini, A.M.; Carrara, E.; et al. Clinical Outcome in Solid Organ Transplant Recipients Affected by COVID-19 Compared to General Population: A Systematic Review and Meta-Analysis. Clin. Microbiol. Infect. 2022, 28, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Oltean, M.; Søfteland, J.M.; Bagge, J.; Ekelund, J.; Felldin, M.; Schult, A.; Magnusson, J.; Friman, V.; Karason, K. COVID-19 in Kidney Transplant Recipients: A Systematic Review of the Case Series Available Three Months into the Pandemic. Infect. Dis. 2020, 52, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Genuardi, M.V.; Moss, N.; Najjar, S.S.; Houston, B.A.; Shore, S.; Vorovich, E.; Atluri, P.; Molina, M.; Chambers, S.; Sharkoski, T.; et al. Coronavirus Disease 2019 in Heart Transplant Recipients: Risk Factors, Immunosuppression, and Outcomes. J. Heart Lung Transplant. 2021, 40, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, Z.S.; Williams, J.D.; Vahia, A.; Fadel, R.; Parraga Acosta, T.; Prashar, R.; Shrivastava, P.; Khoury, N.; Pinto Corrales, J.; Williams, C.; et al. Clinical Characteristics and Outcomes of COVID-19 in Solid Organ Transplant Recipients: A Cohort Study. Am. J. Transplant. 2020, 20, 3051–3060. [Google Scholar] [CrossRef] [PubMed]
- Chavarot, N.; Gueguen, J.; Bonnet, G.; Jdidou, M.; Trimaille, A.; Burger, C.; Amrouche, L.; Weizman, O.; Pommier, T.; Aubert, O.; et al. COVID-19 Severity in Kidney Transplant Recipients Is Similar to Nontransplant Patients with Similar Comorbidities. Am. J. Transplant. 2021, 21, 1285–1294. [Google Scholar] [CrossRef]
- Kamp, J.C.; Hinrichs, J.B.; Fuge, J.; Ewen, R.; Gottlieb, J. COVID-19 in Lung Transplant Recipients—Risk Prediction and Outcomes. PLoS ONE 2021, 16, e0257807. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.V.; Tevethia, H.V.; Premkumar, M.; Arab, J.P.; Candia, R.; Kumar, K.; Kumar, P.; Sharma, M.; Rao, P.N.; Reddy, D.N. Impact of COVID-19 on Liver Transplant Recipients–A Systematic Review and Meta-Analysis. EClinicalMedicine 2021, 38, 101025. [Google Scholar] [CrossRef]
- Sharma, P.; Chen, V.; Fung, C.M.; Troost, J.P.; Patel, V.N.; Combs, M.; Norman, S.; Garg, P.; Colvin, M.; Aaronson, K.; et al. COVID-19 Outcomes among Solid Organ Transplant Recipients: A Case-Control Study. Transplantation 2021, 105, 128–137. [Google Scholar] [CrossRef]
- Bojesen, A.B.; Lund, A.; Mortensen, F.V.; Kirkegård, J. Splenectomy and Risk of COVID-19 Infection, Hospitalisation, and Death. Infect. Dis. 2021, 53, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ni, S.-Y.; Yan, W.; Lu, Q.-D.; Zhao, Y.-M.; Xu, Y.-Y.; Mei, H.; Shi, L.; Yuan, K.; Han, Y.; et al. Mental and Neurological Disorders and Risk of COVID-19 Susceptibility, Illness Severity and Mortality: A Systematic Review, Meta-Analysis and Call for Action. EClinicalMedicine 2021, 40, 101111. [Google Scholar] [CrossRef] [PubMed]
- Masoli, J.; Kuo, C.L.; Atkins, J.; Pilling, L.; Delgado, J.; Kuchel, G.; Melzer, D. 38 Dementia, Apoe and COVID-19 Severity. Age Ageing 2021, 50, i7–i11. [Google Scholar] [CrossRef]
- Hariyanto, T.I.; Putri, C.; Arisa, J.; Situmeang, R.F.V.; Kurniawan, A. Dementia and Outcomes from Coronavirus Disease 2019 (COVID-19) Pneumonia: A Systematic Review and Meta-Analysis. Arch. Gerontol. Geriatr. 2021, 93, 104299. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Travaglio, M.; Popovic, R.; Leal, N.S.; Martins, L.M. Alzheimer’s and Parkinson’s Diseases Predict Different COVID-19 Outcomes: A UK Biobank Study. Geriatrics 2021, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, C.; Sun, Y.; Huang, W.; Ye, K. Cognitive Disorders Associated with Hospitalization of COVID-19: Results from an Observational Cohort Study. Brain. Behav. Immun. 2021, 91, 383–392. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Ren, L.; Shao, Y.; Tao, W.; Dai, X. Preexisting Mental Disorders Increase the Risk of COVID-19 Infection and Associated Mortality. Front. Public Health 2021, 9, 684112. [Google Scholar] [CrossRef]
- Zuin, M.; Guasti, P.; Roncon, L.; Cervellati, C.; Zuliani, G. Dementia and the Risk of Death in Elderly Patients with COVID-19 Infection: Systematic Review and Meta-Analysis. Int. J. Geriatr. Psychiatry 2021, 36, 697–703. [Google Scholar] [CrossRef]
- Bellou, V.; Tzoulaki, I.; Smeden, M.V.; Moons, K.G.M.; Evangelou, E.; Belbasis, L. Prognostic Factors for Adverse Outcomes in Patients with COVID-19: A Field-Wide Systematic Review and Meta-Analysis. Eur. Respir. J. 2022, 59, 2002964. [Google Scholar] [CrossRef]
- Liu, N.; Sun, J.; Wang, X.; Zhao, M.; Huang, Q.; Li, H. The Impact of Dementia on the Clinical Outcome of COVID-19: A Systematic Review and Meta-Analysis. J. Alzheimers Dis. 2020, 78, 1775–1782. [Google Scholar] [CrossRef]
- Filardo, T.D.; Khan, M.R.; Krawczyk, N.; Galitzer, H.; Karmen-Tuohy, S.; Coffee, M.; Schaye, V.E.; Eckhardt, B.J.; Cohen, G.M. Comorbidity and Clinical Factors Associated with COVID-19 Critical Illness and Mortality at a Large Public Hospital in New York City in the Early Phase of the Pandemic (March–April 2020). PLoS ONE 2020, 15, e0242760. [Google Scholar] [CrossRef] [PubMed]
- Samuels, S.; Niu, J.; Sareli, C.; Eckardt, P. The Epidemiology and Predictors of Outcomes among Confirmed COVID-19 Cases in a Large Community Healthcare System in South Florida. J. Community Health 2021, 46, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Chang, I.B.; Kim, Y.H.; Min, C.Y.; Yoo, D.M.; Choi, H.G. The Association of Pre-Existing Diagnoses of Alzheimer’s Disease and Parkinson’s Disease and Coronavirus Disease 2019 Infection, Severity and Mortality: Results from the Korean National Health Insurance Database. Front. Aging Neurosci. 2022, 14, 821235. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.J.; Chang, Y.; Jeon, J.; Shin, J.I.; Song, T.-J.; Kim, J. Association of Alzheimer’s Disease with COVID-19 Susceptibility and Severe Complications: A Nationwide Cohort Study. J. Alzheimers Dis. 2022, 87, 701–710. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Kazis, L.E.; Xia, W. Clinical Outcomes of COVID-19 Infection among Patients with Alzheimer’s Disease or Mild Cognitive Impairment. Alzheimers Dement. 2022, 18, 911–923. [Google Scholar] [CrossRef]
- Honardoost, M.; Janani, L.; Aghili, R.; Emami, Z.; Khamseh, M.E. The Association between Presence of Comorbidities and COVID-19 Severity: A Systematic Review and Meta-Analysis. Cerebrovasc. Dis. 2021, 50, 132–140. [Google Scholar] [CrossRef]
- Yu, J.-N.; Wu, B.-B.; Yang, J.; Lei, X.-L.; Shen, W.-Q. Cardio-Cerebrovascular Disease Is Associated with Severity and Mortality of COVID-19: A Systematic Review and Meta-Analysis. Biol. Res. Nurs. 2021, 23, 258–269. [Google Scholar] [CrossRef]
- Siepmann, T.; Sedghi, A.; Barlinn, J.; de With, K.; Mirow, L.; Wolz, M.; Gruenewald, T.; Helbig, S.; Schroettner, P.; Winzer, S.; et al. Association of History of Cerebrovascular Disease with Severity of COVID-19. J. Neurol. 2021, 268, 773–784. [Google Scholar] [CrossRef]
- Pranata, R.; Huang, I.; Lim, M.A.; Wahjoepramono, E.J.; July, J. Impact of Cerebrovascular and Cardiovascular Diseases on Mortality and Severity of COVID-19–Systematic Review, Meta-Analysis, and Meta-Regression. J. Stroke Cerebrovasc. Dis. 2020, 29, 104949. [Google Scholar] [CrossRef]
- Ramphul, K.; Lohana, P.; Ramphul, Y.; Park, Y.; Mejias, S.; Dhillon, B.K.; Sombans, S.; Verma, R. Hypertension, Diabetes Mellitus, and Cerebrovascular Disease Predispose to a More Severe Outcome of COVID-19. Arch. Med. Sci. Atheroscler. Dis. 2021, 6, e30–e39. [Google Scholar] [CrossRef]
- Patel, U.; Malik, P.; Shah, D.; Patel, A.; Dhamoon, M.; Jani, V. Pre-Existing Cerebrovascular Disease and Poor Outcomes of COVID-19 Hospitalized Patients: A Meta-Analysis. J. Neurol. 2021, 268, 240–247. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, W.; Wang, Y.; Wang, X.; Liu, Y.; Zhao, S.; Long, D.; Chen, L.; Yu, L. Clinical Course and Mortality of Stroke Patients with Coronavirus Disease 2019 in Wuhan, China. Stroke 2020, 51, 2674–2682. [Google Scholar] [CrossRef]
- Kummer, B.R.; Klang, E.; Stein, L.K.; Dhamoon, M.S.; Jetté, N. History of Stroke Is Independently Associated with In-Hospital Death in Patients with COVID-19. Stroke 2020, 51, 3112–3114. [Google Scholar] [CrossRef]
- Eskandar, E.N.; Altschul, D.J.; de la Garza Ramos, R.; Cezayirli, P.; Unda, S.R.; Benton, J.; Dardick, J.; Toma, A.; Patel, N.; Malaviya, A.; et al. Neurologic Syndromes Predict Higher In-Hospital Mortality in COVID-19. Neurology 2021, 96, e1527–e1538. [Google Scholar] [CrossRef]
- Yoo, J.; Kim, J.H.; Jeon, J.; Kim, J.; Song, T.-J. Risk of COVID-19 Infection and of Severe Complications among People with Epilepsy: A Nationwide Cohort Study. Neurology 2022, 98, e1886–e1892. [Google Scholar] [CrossRef]
- Hariyanto, T.I.; Kurniawan, A. Obstructive Sleep Apnea (OSA) and Outcomes from Coronavirus Disease 2019 (COVID-19) Pneumonia: A Systematic Review and Meta-Analysis. Sleep Med. 2021, 82, 47–53. [Google Scholar] [CrossRef]
- Rögnvaldsson, K.G.; Eyþórsson, E.S.; Emilsson, Ö.I.; Eysteinsdóttir, B.; Pálsson, R.; Gottfreðsson, M.; Guðmundsson, G.; Steingrímsson, V. Obstructive Sleep Apnea Is an Independent Risk Factor for Severe COVID-19: A Population-Based Study. Sleep 2022, 45, zsab272. [Google Scholar] [CrossRef]
- Chung, F.; Waseem, R.; Pham, C.; Penzel, T.; Han, F.; Bjorvatn, B.; Morin, C.M.; Holzinger, B.; Espie, C.A.; Benedict, C.; et al. The Association between High Risk of Sleep Apnea, Comorbidities, and Risk of COVID-19: A Population-Based International Harmonized Study. Sleep Breath. 2021, 25, 849–860. [Google Scholar] [CrossRef]
- Hu, M.; Han, X.; Ren, J.; Wang, Y.; Yang, H. Significant Association of Obstructive Sleep Apnoea with Increased Risk for Fatal COVID-19: A Quantitative Meta-Analysis Based on Adjusted Effect Estimates. Sleep Med. Rev. 2022, 63, 101624. [Google Scholar] [CrossRef]
- Goldstein, C.A.; Rizvydeen, M.; Conroy, D.A.; O’Brien Louise, M.; Gupta, G.; Somers, E.C.; Sharma, P.; Golob, J.L.; Troost, J.P.; Burgess, H.J. The Prevalence and Impact of Pre-Existing Sleep Disorder Diagnoses and Objective Sleep Parameters in Patients Hospitalized for COVID-19. J. Clin. Sleep Med. 2021, 17, 1039–1050. [Google Scholar] [CrossRef]
- Vignatelli, L.; Zenesini, C.; Belotti, L.M.B.; Baldin, E.; Bonavina, G.; Calandra-Buonaura, G.; Cortelli, P.; Descovich, C.; Fabbri, G.; Giannini, G.; et al. Risk of Hospitalization and Death for COVID-19 in People with Parkinson’s Disease or Parkinsonism. Mov. Disord. 2021, 36, 1–10. [Google Scholar] [CrossRef]
- El-Qushayri, A.E.; Ghozy, S.; Reda, A.; Kamel, A.M.A.; Abbas, A.S.; Dmytriw, A.A. The Impact of Parkinson’s Disease on Manifestations and Outcomes of COVID-19 Patients: A Systematic Review and Meta-Analysis. Rev. Med. Virol. 2022, 32, e2278. [Google Scholar] [CrossRef] [PubMed]
- Ceban, F.; Nogo, D.; Carvalho, I.P.; Lee, Y.; Nasri, F.; Xiong, J.; Lui, L.M.W.; Subramaniapillai, M.; Gill, H.; Liu, R.N.; et al. Association Between Mood Disorders and Risk of COVID-19 Infection, Hospitalization, and Death: A Systematic Review and Meta-Analysis. JAMA Psychiatry 2021, 78, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Vai, B.; Mazza, M.G.; Delli Colli, C.; Foiselle, M.; Allen, B.; Benedetti, F.; Borsini, A.; Casanova Dias, M.; Tamouza, R.; Leboyer, M.; et al. Mental Disorders and Risk of COVID-19-Related Mortality, Hospitalisation, and Intensive Care Unit Admission: A Systematic Review and Meta-Analysis. Lancet Psychiatry 2021, 8, 797–812. [Google Scholar] [CrossRef] [PubMed]
- Castro, V.M.; Gunning, F.M.; McCoy, T.H.; Perlis, R.H. Mood Disorders and Outcomes of COVID-19 Hospitalizations. Am. J. Psychiatry 2021, 178, 541–547. [Google Scholar] [CrossRef]
- Egede, C.; Dawson, A.Z.; Walker, R.J.; Garacci, E.; Campbell, J.A.; Egede, L.E. Relationship between Mental Health Diagnoses and COVID-19 Test Positivity, Hospitalization, and Mortality in Southeast Wisconsin. Psychol. Med. 2021, 1–9. [Google Scholar] [CrossRef]
- Barcella, C.A.; Polcwiartek, C.; Mohr, G.H.; Hodges, G.; Søndergaard, K.; Niels Bang, C.; Andersen, M.P.; Fosbøl, E.; Køber, L.; Schou, M.; et al. Severe Mental Illness Is Associated with Increased Mortality and Severe Course of COVID-19. Acta Psychiatr. Scand. 2021, 144, 82–91. [Google Scholar] [CrossRef]
- Fond, G.; Pauly, V.; Leone, M.; Orleans, V.; Garosi, A.; Lancon, C.; Auquier, P.; Baumstarck, K.; Llorca, P.-M.; Boyer, L. Mortality among Inpatients with Bipolar Disorders and COVID-19: A Propensity Score Matching Analysis in a National French Cohort Study. Psychol. Med. 2021, 1–10. [Google Scholar] [CrossRef]
- Yang, H.; Chen, W.; Hu, Y.; Chen, Y.; Zeng, Y.; Sun, Y.; Ying, Z.; He, J.; Qu, Y.; Lu, D.; et al. Pre-Pandemic Psychiatric Disorders and Risk of COVID-19: A UK Biobank Cohort Analysis. Lancet Healthy Longev. 2020, 1, e69–e79. [Google Scholar] [CrossRef]
- Wang, S.; Quan, L.; Ding, M.; Kang, J.H.; Koenen, K.C.; Kubzansky, L.D.; Branch-Elliman, W.; Chavarro, J.E.; Roberts, A.L. Depression, Worry, and Loneliness Are Associated with Subsequent Risk of Hospitalization for COVID-19: A Prospective Study. Psychol. Med. 2022, 1–10. [Google Scholar] [CrossRef]
- Merzon, E.; Weiss, M.D.; Cortese, S.; Rotem, A.; Schneider, T.; Craig, S.G.; Vinker, S.; Golan Cohen, A.; Green, I.; Ashkenazi, S.; et al. The Association between ADHD and the Severity of COVID-19 Infection. J. Atten. Disord. 2022, 26, 491–501. [Google Scholar] [CrossRef]
- Velásquez García, H.A.; Wilton, J.; Smolina, K.; Chong, M.; Rasali, D.; Otterstatter, M.; Rose, C.; Prystajecky, N.; David, S.; Galanis, E.; et al. Mental Health and Substance Use Associated with Hospitalization among People with COVID-19: A Population-Based Cohort Study. Viruses 2021, 13, 2196. [Google Scholar] [CrossRef]
- Nishimi, K.; Neylan, T.C.; Bertenthal, D.; Dolsen, E.A.; Seal, K.H.; O’Donovan, A. Post-Traumatic Stress Disorder and Risk for Hospitalization and Death Following COVID-19 Infection. Transl. Psychiatry 2022, 12, 482. [Google Scholar] [CrossRef]
- Fond, G.; Pauly, V.; Leone, M.; Llorca, P.-M.; Orleans, V.; Loundou, A.; Lancon, C.; Auquier, P.; Baumstarck, K.; Boyer, L. Disparities in Intensive Care Unit Admission and Mortality among Patients with Schizophrenia and COVID-19: A National Cohort Study. Schizophr. Bull. 2021, 47, 624–634. [Google Scholar] [CrossRef]
- Nemani, K.; Li, C.; Olfson, M.; Blessing, E.M.; Razavian, N.; Chen, J.; Petkova, E.; Goff, D.C. Association of Psychiatric Disorders with Mortality among Patients with COVID-19. JAMA Psychiatry 2021, 78, 380–386. [Google Scholar] [CrossRef]
- Pardamean, E.; Roan, W.; Iskandar, K.T.A.; Prayangga, R.; Hariyanto, T.I. Mortality from Coronavirus Disease 2019 (COVID-19) in Patients with Schizophrenia: A Systematic Review, Meta-Analysis and Meta-Regression. Gen. Hosp. Psychiatry 2022, 75, 61–67. [Google Scholar] [CrossRef]
- Baillargeon, J.; Polychronopoulou, E.; Kuo, Y.-F.; Raji, M.A. The Impact of Substance Use Disorder on COVID-19 Outcomes. Psychiatr. Serv. 2021, 72, 578–581. [Google Scholar] [CrossRef]
- Hasin, D.S.; Fink, D.S.; Olfson, M.; Saxon, A.J.; Malte, C.; Keyes, K.M.; Gradus, J.L.; Cerdá, M.; Maynard, C.C.; Keyhani, S.; et al. Substance Use Disorders and COVID-19: An Analysis of Nation-Wide Veterans Health Administration Electronic Health Records. Drug Alcohol Depend. 2022, 234, 109383. [Google Scholar] [CrossRef]
- Lefere, S.; Tacke, F. Macrophages in Obesity and Non-Alcoholic Fatty Liver Disease: Crosstalk with Metabolism. JHEP Rep. 2019, 1, 30–43. [Google Scholar] [CrossRef]
- Da, B.L.; Im, G.Y.; Schiano, T.D. Coronavirus Disease 2019 Hangover: A Rising Tide of Alcohol Use Disorder and Alcohol-Associated Liver Disease. Hepatology 2020, 72, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ni, C.; Gao, R.; Wang, Y.; Yang, L.; Wei, J.; Lv, T.; Liang, J.; Zhang, Q.; Xu, W.; et al. Recapitulation of SARS-CoV-2 Infection and Cholangiocyte Damage with Human Liver Ductal Organoids. Protein Cell 2020, 11, 771–775. [Google Scholar] [CrossRef] [Green Version]
- Parveen, R.; Sehar, N.; Bajpai, R.; Agarwal, N.B. Association of Diabetes and Hypertension with Disease Severity in COVID-19 Patients: A Systematic Literature Review and Exploratory Meta-Analysis. Diabetes Res. Clin. Pract. 2020, 166, 108295. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhou, F.; Sun, W.; Chen, L.; Lan, L.; Li, H.; Xiao, F.; Li, Y.; Kolachalama, V.B.; Li, Y.; et al. Relationship Between Serum Severe Acute Respiratory Syndrome Coronavirus 2 Nucleic Acid and Organ Damage in Coronavirus 2019 Patients: A Cohort Study. Clin. Infect. Dis. 2021, 73, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Huang, D.; Yu, H.; Zhu, Z.; Xia, Z.; Su, Y.; Li, Z.; Zhou, G.; Gou, J.; Qu, J.; et al. COVID-19: Abnormal Liver Function Tests. J. Hepatol. 2020, 73, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Finn, O.J. Immuno-Oncology: Understanding the Function and Dysfunction of the Immune System in Cancer. Ann. Oncol. 2012, 23, viii6–viii9. [Google Scholar] [CrossRef] [PubMed]
- Ménétrier-Caux, C.; Ray-Coquard, I.; Blay, J.-Y.; Caux, C. Lymphopenia in Cancer Patients and Its Effects on Response to Immunotherapy: An Opportunity for Combination with Cytokines? J. Immunother. Cancer 2019, 7, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamilos, G.; Lionakis, M.S.; Kontoyiannis, D.P. Are All Patients with Cancer at Heightened Risk for Severe Coronavirus Disease 2019 (COVID-19)? Clin. Infect. Dis. 2021, 72, 351–356. [Google Scholar] [CrossRef]
- Di Felice, G.; Visci, G.; Teglia, F.; Angelini, M.; Boffetta, P. Effect of Cancer on Outcome of COVID-19 Patients: A Systematic Review and Meta-Analysis of Studies of Unvaccinated Patients. eLife 2022, 11, e74634. [Google Scholar] [CrossRef]
- Desai, A.; Gupta, R.; Advani, S.; Ouellette, L.; Kuderer, N.M.; Lyman, G.H.; Li, A. Mortality in Hospitalized Patients with Cancer and Coronavirus Disease 2019: A Systematic Review and Meta-Analysis of Cohort Studies. Cancer 2021, 127, 1459–1468. [Google Scholar] [CrossRef]
- Dai, M.; Liu, D.; Liu, M.; Zhou, F.; Li, G.; Chen, Z.; Zhang, Z.; You, H.; Wu, M.; Zheng, Q.; et al. Patients with Cancer Appear More Vulnerable to SARS-CoV-2: A Multicenter Study during the COVID-19 Outbreak. Cancer Discov. 2020, 10, 783–791. [Google Scholar] [CrossRef]
- Basse, C.; Diakite, S.; Servois, V.; Frelaut, M.; Noret, A.; Bellesoeur, A.; Moreau, P.; Massiani, M.-A.; Bouyer, A.-S.; Vuagnat, P.; et al. Characteristics and Outcome of SARS-CoV-2 Infection in Cancer Patients. JNCI Cancer Spectr. 2021, 5, pkaa090. [Google Scholar] [CrossRef]
- Yekedüz, E.; Utkan, G.; Ürün, Y. A Systematic Review and Meta-Analysis: The Effect of Active Cancer Treatment on Severity of COVID-19. Eur. J. Cancer 2020, 141, 92–104. [Google Scholar] [CrossRef]
- Robilotti, E.V.; Babady, N.E.; Mead, P.A.; Rolling, T.; Perez-Johnston, R.; Bernardes, M.; Bogler, Y.; Caldararo, M.; Figueroa, C.J.; Glickman, M.S.; et al. Determinants of COVID-19 Disease Severity in Patients with Cancer. Nat. Med. 2020, 26, 1218–1223. [Google Scholar] [CrossRef]
- Fagni, F.; Simon, D.; Tascilar, K.; Schoenau, V.; Sticherling, M.; Neurath, M.F.; Schett, G. COVID-19 and Immune-Mediated Inflammatory Diseases: Effect of Disease and Treatment on COVID-19 Outcomes and Vaccine Responses. Lancet Rheumatol. 2021, 3, e724–e736. [Google Scholar] [CrossRef]
- Chertok Shacham, E.; Ishay, A. New Insights on Effects of Glucocorticoids in SARS-CoV-2 Infection. Endocr. Pract. 2022, 28, 1100–1106. [Google Scholar] [CrossRef]
- Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [CrossRef]
- Singanayagam, A.; Johnston, S.L. Long-Term Impact of Inhaled Corticosteroid Use in Asthma and Chronic Obstructive Pulmonary Disease (COPD): Review of Mechanisms That Underlie Risks. J. Allergy Clin. Immunol. 2020, 146, 1292–1294. [Google Scholar] [CrossRef] [Green Version]
- Singanayagam, A.; Glanville, N.; Cuthbertson, L.; Bartlett, N.W.; Finney, L.J.; Turek, E.; Bakhsoliani, E.; Calderazzo, M.A.; Trujillo-Torralbo, M.-B.; Footitt, J.; et al. Inhaled Corticosteroid Suppression of Cathelicidin Drives Dysbiosis and Bacterial Infection in Chronic Obstructive Pulmonary Disease. Sci. Transl. Med. 2019, 11, eaav3879. [Google Scholar] [CrossRef]
- Ameratunga, R.; Longhurst, H.; Steele, R.; Lehnert, K.; Leung, E.; Brooks, A.E.S.; Woon, S.-T. Common Variable Immunodeficiency Disorders, T-Cell Responses to SARS-CoV-2 Vaccines, and the Risk of Chronic COVID-19. J. Allergy Clin. Immunol. Pract. 2021, 9, 3575–3583. [Google Scholar] [CrossRef]
- Onisiforou, A.; Spyrou, G.M. Systems Bioinformatics Reveals Possible Relationship between COVID-19 and the Development of Neurological Diseases and Neuropsychiatric Disorders. Viruses 2022, 14, 2270. [Google Scholar] [CrossRef]
- Roy, E.R.; Wang, B.; Wan, Y.; Chiu, G.; Cole, A.; Yin, Z.; Propson, N.E.; Xu, Y.; Jankowsky, J.L.; Liu, Z.; et al. Type I Interferon Response Drives Neuroinflammation and Synapse Loss in Alzheimer Disease. J. Clin. Investig. 2020, 130, 1912–1930. [Google Scholar] [CrossRef]
- Finch, C.E.; Kulminski, A.M. The ApoE Locus and COVID-19: Are We Going Where We Have Been? J. Gerontol. A. Biol. Sci. Med. Sci. 2020, 76, e1–e3. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, Y.; Wang, Y.; Ke, Q.; Cui, L. The COVID-19 Pandemic and Alzheimer’s Disease: Mutual Risks and Mechanisms. Transl. Neurodegener. 2022, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, S.; Saito, H.; Kanda, A.; Nakajoh, M.; Takahashi, H.; Arai, H.; Sasaki, H. Impaired Efficacy of Cough in Patients with Parkinson Disease. Chest 2003, 124, 1009–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dono, F.; Nucera, B.; Lanzone, J.; Evangelista, G.; Rinaldi, F.; Speranza, R.; Troisi, S.; Tinti, L.; Russo, M.; Di Pietro, M.; et al. Status Epilepticus and COVID-19: A Systematic Review. Epilepsy Behav. EB 2021, 118, 107887. [Google Scholar] [CrossRef]
- Romagnolo, A.; Imbalzano, G.; Artusi, C.A.; Balestrino, R.; Ledda, C.; De Rosa, F.G.; Riccardini, F.; Montanaro, E.; Bozzali, M.; Rizzone, M.G.; et al. Neurological Comorbidities and COVID-19-Related Case Fatality: A Cohort Study. J. Neurol. Sci. 2021, 428, 117610. [Google Scholar] [CrossRef]
- Galea, J.; Brough, D. The Role of Inflammation and Interleukin-1 in Acute Cerebrovascular Disease. J. Inflamm. Res. 2013, 6, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Yuan, N.; Chen, Y.; Xia, Y.; Dai, J.; Liu, C. Inflammation-Related Biomarkers in Major Psychiatric Disorders: A Cross-Disorder Assessment of Reproducibility and Specificity in 43 Meta-Analyses. Transl. Psychiatry 2019, 9, 233. [Google Scholar] [CrossRef] [Green Version]
- Cauley, L.S.; Vella, A.T. Why Is Coinfection with Influenza Virus and Bacteria so Difficult to Control? Discov. Med. 2015, 19, 33–40. [Google Scholar]
- Gill, J.R.; Sheng, Z.-M.; Ely, S.F.; Guinee, D.G., Jr.; Beasley, M.B.; Suh, J.; Deshpande, C.; Mollura, D.J.; Morens, D.M.; Bray, M.; et al. Pulmonary Pathologic Findings of Fatal 2009 Pandemic Influenza A/H1N1 Viral Infections. Arch. Pathol. Lab. Med. 2010, 134, 235–243. [Google Scholar] [CrossRef]
- Bisno, A.L.; Griffin, J.P.; Van Epps, K.A.; Niell, H.B.; Rytel, M.W. Pneumonia and Hong Kong Influenza: A Prospective Study of the 1968-1969 Epidemic. Am. J. Med. Sci. 1971, 261, 251–263. [Google Scholar] [CrossRef]
- Omoush, S.A.; Alzyoud, J.A.M. The Prevalence and Impact of Coinfection and Superinfection on the Severity and Outcome of COVID-19 Infection: An Updated Literature Review. Pathogens 2022, 11, 445. [Google Scholar] [CrossRef] [PubMed]
- Musuuza, J.S.; Watson, L.; Parmasad, V.; Putman-Buehler, N.; Christensen, L.; Safdar, N. Prevalence and Outcomes of Co-Infection and Superinfection with SARS-CoV-2 and Other Pathogens: A Systematic Review and Meta-Analysis. PLoS ONE 2021, 16, e0251170. [Google Scholar] [CrossRef]
- Tsheten, T.; Clements, A.C.A.; Gray, D.J.; Adhikary, R.K.; Wangdi, K. Clinical Features and Outcomes of COVID-19 and Dengue Co-Infection: A Systematic Review. BMC Infect. Dis. 2021, 21, 729. [Google Scholar] [CrossRef]
- Simonnet, A.; Engelmann, I.; Moreau, A.-S.; Garcia, B.; Six, S.; El Kalioubie, A.; Robriquet, L.; Hober, D.; Jourdain, M. High Incidence of Epstein-Barr Virus, Cytomegalovirus, and Human-Herpes Virus-6 Reactivations in Critically Ill Patients with COVID-19. Infect. Dis. Now 2021, 51, 296–299. [Google Scholar] [CrossRef]
- Taherifard, E.; Movahed, H.; Kiani Salmi, S.; Taherifard, A.; Abdollahifard, S.; Taherifard, E. Cytomegalovirus Coinfection in Patients with Severe Acute Respiratory Syndrome Coronavirus 2 Infection: A Systematic Review of Reported Cases. Infect. Dis. 2022, 54, 543–557. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, R.; Joshi, S.R.; Misra, A. Mucormycosis in COVID-19: A Systematic Review of Cases Reported Worldwide and in India. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 102146. [Google Scholar] [CrossRef]
- Szydłowicz, M.; Matos, O. Pneumocystis Pneumonia in the COVID-19 Pandemic Era: Similarities and Challenges. Trends Parasitol. 2021, 37, 859–862. [Google Scholar] [CrossRef]
- Hernández-Terán, A.; Mejía-Nepomuceno, F.; Herrera, M.T.; Barreto, O.; García, E.; Castillejos, M.; Boukadida, C.; Matias-Florentino, M.; Rincón-Rubio, A.; Avila-Rios, S.; et al. Dysbiosis and Structural Disruption of the Respiratory Microbiota in COVID-19 Patients with Severe and Fatal Outcomes. Sci. Rep. 2021, 11, 21297. [Google Scholar] [CrossRef]
- Sender, V.; Hentrich, K.; Henriques-Normark, B. Virus-Induced Changes of the Respiratory Tract Environment Promote Secondary Infections with Streptococcus Pneumoniae. Front. Cell. Infect. Microbiol. 2021, 11, 643326. [Google Scholar] [CrossRef]
- de Buhr, N.; von Köckritz-Blickwede, M. The Balance of Neutrophil Extracellular Trap Formation and Nuclease Degradation: An Unknown Role of Bacterial Coinfections in COVID-19 Patients? mBio 2021, 12, e03304-20. [Google Scholar] [CrossRef]
- Rangel, K.; Chagas, T.P.G.; De-Simone, S.G. Acinetobacter Baumannii Infections in Times of COVID-19 Pandemic. Pathogens 2021, 10, 1006. [Google Scholar] [CrossRef]
- Westblade, L.F.; Simon, M.S.; Satlin, M.J. Bacterial Coinfections in Coronavirus Disease 2019. Trends Microbiol. 2021, 29, 930–941. [Google Scholar] [CrossRef]
- Silva, D.L.; Lima, C.M.; Magalhães, V.C.R.; Baltazar, L.M.; Peres, N.T.A.; Caligiorne, R.B.; Moura, A.S.; Fereguetti, T.; Martins, J.C.; Rabelo, L.F.; et al. Fungal and Bacterial Coinfections Increase Mortality of Severely Ill COVID-19 Patients. J. Hosp. Infect. 2021, 113, 145–154. [Google Scholar] [CrossRef]
- Amin, D.; McKitish, K.; Shah, P.S. Association of Mortality and Recent Mycoplasma Pneumoniae Infection in COVID-19 Patients. J. Med. Virol. 2021, 93, 1180–1183. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Sanjuan, G.; Moreno-García, E.; Puerta-Alcalde, P.; Garcia-Pouton, N.; Chumbita, M.; Fernandez-Pittol, M.; Pitart, C.; Inciarte, A.; Bodro, M.; et al. Incidence of Co-Infections and Superinfections in Hospitalized Patients with COVID-19: A Retrospective Cohort Study. Clin. Microbiol. Infect. 2021, 27, 83. [Google Scholar] [CrossRef]
- Hedberg, P.; Johansson, N.; Ternhag, A.; Abdel-Halim, L.; Hedlund, J.; Nauclér, P. Bacterial Co-Infections in Community-Acquired Pneumonia Caused by SARS-CoV-2, Influenza Virus and Respiratory Syncytial Virus. BMC Infect. Dis. 2022, 22, 108. [Google Scholar] [CrossRef]
- Falcone, M.; Tiseo, G.; Giordano, C.; Leonildi, A.; Menichini, M.; Vecchione, A.; Pistello, M.; Guarracino, F.; Ghiadoni, L.; Forfori, F.; et al. Predictors of Hospital-Acquired Bacterial and Fungal Superinfections in COVID-19: A Prospective Observational Study. J. Antimicrob. Chemother. 2021, 76, 1078–1084. [Google Scholar] [CrossRef]
- Wang, L.; Amin, A.K.; Khanna, P.; Aali, A.; McGregor, A.; Bassett, P.; Gopal Rao, G. An Observational Cohort Study of Bacterial Co-Infection and Implications for Empirical Antibiotic Therapy in Patients Presenting with COVID-19 to Hospitals in North West London. J. Antimicrob. Chemother. 2021, 76, 796–803. [Google Scholar] [CrossRef]
- Mitsi, E.; Reiné, J.; Urban, B.C.; Solórzano, C.; Nikolaou, E.; Hyder-Wright, A.D.; Pojar, S.; Howard, A.; Hitchins, L.; Glynn, S.; et al. Streptococcus Pneumoniae Colonization Associates with Impaired Adaptive Immune Responses against SARS-CoV-2. J. Clin. Investig. 2022, 132, e157124. [Google Scholar] [CrossRef]
- SARS-CoV-2, Bacterial Co-Infections, and AMR: The Deadly Trio in COVID-19? EMBO Mol. Med. 2020, 12, e12560. [CrossRef]
- Duployez, C.; Guern, R.L.; Tinez, C.; Lejeune, A.-L.; Robriquet, L.; Six, S.; Loïez, C.; Wallet, F. Panton-Valentine Leukocidin–Secreting Staphylococcus Aureus Pneumonia Complicating COVID-19. Emerg. Infect. Dis. 2020, 26, 1939. [Google Scholar] [CrossRef]
- Nieto-Moro, M.; Ecclesia, F.G.; Tomé-Masa, I.; De Lama Caro-Patón, G.; Leoz-Gordillo, I.; Cabrero-Hernández, M.; García-Salido, A. SARS-CoV-2 and Streptococcus Pneumoniae Coinfection as a Cause of Severe Pneumonia in an Infant. Pediatr. Pulmonol. 2020, 55, 2198–2200. [Google Scholar] [CrossRef]
- Riccò, M.; Ferraro, P.; Peruzzi, S.; Zaniboni, A.; Ranzieri, S. SARS-CoV-2–Legionella Co-Infections: A Systematic Review and Meta-Analysis (2020–2021). Microorganisms 2022, 10, 499. [Google Scholar] [CrossRef]
- Shah, T.; Shah, Z.; Yasmeen, N.; Baloch, Z.; Xia, X. Pathogenesis of SARS-CoV-2 and Mycobacterium Tuberculosis Coinfection. Front. Immunol. 2022, 13, 909011. [Google Scholar] [CrossRef]
- Sarkar, S.; Khanna, P.; Singh, A.K. Impact of COVID-19 in Patients with Concurrent Co-Infections: A Systematic Review and Meta-Analyses. J. Med. Virol. 2021, 93, 2385–2395. [Google Scholar] [CrossRef]
- Sy, K.T.L.; Haw, N.J.L.; Uy, J. Previous and Active Tuberculosis Increases Risk of Death and Prolongs Recovery in Patients with COVID-19. Infect. Dis. 2020, 52, 902–907. [Google Scholar] [CrossRef]
- Western Cape Department of Health in collaboration with the National Institute for Communicable Diseases, South Africa; Boulle, A.; Davies, M.-A.; Hussey, H.; Ismail, M.; Morden, E.; Vundle, Z.; Zweigenthal, V.; Mahomed, H.; Paleker, M.; et al. Risk Factors for Coronavirus Disease 2019 (COVID-19) Death in a Population Cohort Study from the Western Cape Province, South Africa. Clin. Infect. Dis. 2021, 73, e2005–e2015. [Google Scholar] [CrossRef]
- Mejia, O.R.; Gloag, E.S.; Li, J.; Ruane-Foster, M.; Claeys, T.A.; Farkas, D.; Wang, S.-H.; Farkas, L.; Xin, G.; Robinson, R.T. Mice Infected with Mycobacterium Tuberculosis Are Resistant to Acute Disease Caused by Secondary Infection with SARS-CoV-2. PLOS Pathog. 2022, 18, e1010093. [Google Scholar] [CrossRef]
- TB/COVID-19 Global Study Group. Tuberculosis and COVID-19 Co-Infection: Description of the Global Cohort. Eur. Respir. J. 2022, 59, 2102538. [Google Scholar] [CrossRef]
- Starshinova, A.A.; Kudryavtsev, I.; Malkova, A.; Zinchenko, U.; Karev, V.; Kudlay, D.; Glushkova, A.; Starshinova, A.Y.; Dominguez, J.; Villar-Hernández, R.; et al. Molecular and Cellular Mechanisms of M. Tuberculosis and SARS-CoV-2 Infections—Unexpected Similarities of Pathogenesis and What to Expect from Co-Infection. Int. J. Mol. Sci. 2022, 23, 2235. [Google Scholar] [CrossRef]
- Acharya, D.; Liu, G.; Gack, M.U. Dysregulation of Type I Interferon Responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 397–398. [Google Scholar] [CrossRef]
- Cliff, J.M.; Kaufmann, S.H.E.; McShane, H.; van Helden, P.; O’Garra, A. The Human Immune Response to Tuberculosis and Its Treatment: A View from the Blood. Immunol. Rev. 2015, 264, 88–102. [Google Scholar] [CrossRef] [Green Version]
- Riou, C.; du Bruyn, E.; Stek, C.; Daroowala, R.; Goliath, R.T.; Abrahams, F.; Said-Hartley, Q.; Allwood, B.W.; Hsiao, N.-Y.; Wilkinson, K.A.; et al. Relationship of SARS-CoV-2–Specific CD4 Response to COVID-19 Severity and Impact of HIV-1 and Tuberculosis Coinfection. J. Clin. Investig. 2021, 131, e149125. [Google Scholar] [CrossRef]
- Petrone, L.; Petruccioli, E.; Vanini, V.; Cuzzi, G.; Gualano, G.; Vittozzi, P.; Nicastri, E.; Maffongelli, G.; Grifoni, A.; Sette, A.; et al. Coinfection of Tuberculosis and COVID-19 Limits the Ability to in Vitro Respond to SARS-CoV-2. Int. J. Infect. Dis. 2021, 113, S82–S87. [Google Scholar] [CrossRef]
- Wong, G.L.-H.; Wong, V.W.-S.; Yuen, B.W.-Y.; Tse, Y.-K.; Yip, T.C.-F.; Luk, H.W.-S.; Lui, G.C.-Y.; Chan, H.L.-Y. Risk of Hepatitis B Surface Antigen Seroreversion after Corticosteroid Treatment in Patients with Previous Hepatitis B Virus Exposure. J. Hepatol. 2020, 72, 57–66. [Google Scholar] [CrossRef]
- Yu, R.; Tan, S.; Dan, Y.; Lu, Y.; Zhang, J.; Tan, Z.; He, X.; Xiang, X.; Zhou, Y.; Guo, Y.; et al. Effect of SARS-CoV-2 Coinfection Was Not Apparent on the Dynamics of Chronic Hepatitis B Infection. Virology 2021, 553, 131–134. [Google Scholar] [CrossRef]
- Yang, S.; Wang, S.; Du, M.; Liu, M.; Liu, Y.; He, Y. Patients with COVID-19 and HBV Coinfection Are at Risk of Poor Prognosis. Infect. Dis. Ther. 2022, 11, 1229–1242. [Google Scholar] [CrossRef]
- Kang, S.H.; Cho, D.-H.; Choi, J.; Baik, S.K.; Gwon, J.G.; Kim, M.Y. Association between Chronic Hepatitis B Infection and COVID-19 Outcomes: A Korean Nationwide Cohort Study. PLoS ONE 2021, 16, e0258229. [Google Scholar] [CrossRef]
- Zou, X.; Fang, M.; Li, S.; Wu, L.; Gao, B.; Gao, H.; Ran, X.; Bian, Y.; Li, R.; Yu, S.; et al. Characteristics of Liver Function in Patients with SARS-CoV-2 and Chronic HBV Coinfection. Clin. Gastroenterol. Hepatol. 2021, 19, 597–603. [Google Scholar] [CrossRef]
- Lin, Y.; Yuan, J.; Long, Q.; Hu, J.; Deng, H.; Zhao, Z.; Chen, J.; Lu, M.; Huang, A. Patients with SARS-CoV-2 and HBV Co-Infection Are at Risk of Greater Liver Injury. Genes Dis. 2021, 8, 484–492. [Google Scholar] [CrossRef]
- Ali, N. Relationship Between COVID-19 Infection and Liver Injury: A Review of Recent Data. Front. Med. 2020, 7, 458. [Google Scholar] [CrossRef] [PubMed]
- Xiang, T.-D.; Zheng, X. Interaction between Hepatitis B Virus and SARS-CoV-2 Infections. World J. Gastroenterol. 2021, 27, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Alothaid, H.; Aldughaim, M.S.K.; El Bakkouri, K.; AlMashhadi, S.; Al-Qahtani, A.A. Similarities between the Effect of SARS-CoV-2 and HCV on the Cellular Level, and the Possible Role of Ion Channels in COVID19 Progression: A Review of Potential Targets for Diagnosis and Treatment. Channels 2020, 14, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.A.; Yan, P.; Chotani, R.A.; Shaikh, O.S. Mortality Is Not Increased in SARS-CoV-2 Infected Persons with Hepatitis C Virus Infection. Liver Int. 2021, 41, 1824–1831. [Google Scholar] [CrossRef]
- Rehermann, B. Hepatitis C Virus versus Innate and Adaptive Immune Responses: A Tale of Coevolution and Coexistence. J. Clin. Investig. 2009, 119, 1745–1754. [Google Scholar] [CrossRef] [Green Version]
- Loftis, J.M.; Huckans, M.; Ruimy, S.; Hinrichs, D.J.; Hauser, P. Depressive Symptoms in Patients with Chronic Hepatitis C Are Correlated with Elevated Plasma Levels of Interleukin-1β and Tumor Necrosis Factor-α. Neurosci. Lett. 2008, 430, 264–268. [Google Scholar] [CrossRef] [Green Version]
- Shirley, K.; Loftis, J.M. A Spotlight on HCV and SARS-CoV-2 Co-Infection and Brain Function. Pharmacol. Biochem. Behav. 2022, 217, 173403. [Google Scholar] [CrossRef]
- Gill, K.; Ghazinian, H.; Manch, R.; Gish, R. Hepatitis C Virus as a Systemic Disease: Reaching beyond the Liver. Hepatol. Int. 2016, 10, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Afify, S.; Eysa, B.; Hamid, F.A.; Abo-Elazm, O.M.; Edris, M.A.; Maher, R.; Abdelhalim, A.; Abdel Ghaffar, M.M.; Omran, D.A.; Shousha, H.I. Survival and Outcomes for Co-Infection of Chronic Hepatitis C with and without Cirrhosis and COVID-19: A Multicenter Retrospective Study. World J. Gastroenterol. 2021, 27, 7362–7375. [Google Scholar] [CrossRef]
- Lambarey, H.; Blumenthal, M.J.; Chetram, A.; Joyimbana, W.; Jennings, L.; Tincho, M.B.; Burgers, W.A.; Orrell, C.; Schäfer, G. SARS-CoV-2 Infection Is Associated with Uncontrolled HIV Viral Load in Non-Hospitalized HIV-Infected Patients from Gugulethu, South Africa. Viruses 2022, 14, 1222. [Google Scholar] [CrossRef]
- Moradi, Y.; Soheili, M.; Dehghanbanadaki, H.; Moradi, G.; Moradpour, F.; Mortazavi, S.M.M.; Kohan, H.G.; Zareie, M. The Effect of HIV/AIDS Infection on the Clinical Outcomes of COVID-19: A Meta-Analysis. J. Pharm. Pharm. Sci. 2022, 25, 183–192. [Google Scholar] [CrossRef]
- Danwang, C.; Noubiap, J.J.; Robert, A.; Yombi, J.C. Outcomes of Patients with HIV and COVID-19 Co-Infection: A Systematic Review and Meta-Analysis. AIDS Res. Ther. 2022, 19, 3. [Google Scholar] [CrossRef]
- Yang, X.; Sun, J.; Patel, R.C.; Zhang, J.; Guo, S.; Zheng, Q.; Olex, A.L.; Olatosi, B.; Weissman, S.B.; Islam, J.Y.; et al. Associations between HIV Infection and Clinical Spectrum of COVID-19: A Population Level Analysis Based on US National COVID Cohort Collaborative (N3C) Data. Lancet HIV 2021, 8, e690–e700. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Rentsch, C.T.; MacKenna, B.; Schultze, A.; Mehrkar, A.; Bates, C.J.; Eggo, R.M.; Morton, C.E.; Bacon, S.C.J.; Inglesby, P.; et al. HIV Infection and COVID-19 Death: A Population-Based Cohort Analysis of UK Primary Care Data and Linked National Death Registrations within the OpenSAFELY Platform. Lancet HIV 2021, 8, e24–e32. [Google Scholar] [CrossRef]
- Hoffmann, C.; Casado, J.L.; Härter, G.; Vizcarra, P.; Moreno, A.; Cattaneo, D.; Meraviglia, P.; Spinner, C.D.; Schabaz, F.; Grunwald, S.; et al. Immune Deficiency Is a Risk Factor for Severe COVID-19 in People Living with HIV. HIV Med. 2021, 22, 372–378. [Google Scholar] [CrossRef]
- Ssentongo, P.; Heilbrunn, E.S.; Ssentongo, A.E.; Advani, S.; Chinchilli, V.M.; Nunez, J.J.; Du, P. Epidemiology and Outcomes of COVID-19 in HIV-Infected Individuals: A Systematic Review and Meta-Analysis. Sci. Rep. 2021, 11, 6283. [Google Scholar] [CrossRef]
- Tesoriero, J.M.; Swain, C.-A.E.; Pierce, J.L.; Zamboni, L.; Wu, M.; Holtgrave, D.R.; Gonzalez, C.J.; Udo, T.; Morne, J.E.; Hart-Malloy, R.; et al. COVID-19 Outcomes among Persons Living with or without Diagnosed HIV Infection in New York State. JAMA Netw. Open 2021, 4, e2037069. [Google Scholar] [CrossRef]
- Nomah, D.K.; Reyes-Urueña, J.; Díaz, Y.; Moreno, S.; Aceiton, J.; Bruguera, A.; Vivanco-Hidalgo, R.M.; Llibre, J.M.; Domingo, P.; Falcó, V.; et al. Sociodemographic, Clinical, and Immunological Factors Associated with SARS-CoV-2 Diagnosis and Severe COVID-19 Outcomes in People Living with HIV: A Retrospective Cohort Study. Lancet HIV 2021, 8, e701–e710. [Google Scholar] [CrossRef]
- Mirzaei, H.; McFarland, W.; Karamouzian, M.; Sharifi, H. COVID-19 among People Living with HIV: A Systematic Review. AIDS Behav. 2021, 25, 85–92. [Google Scholar] [CrossRef]
- Sharov, K.S. HIV/SARS-CoV-2 Co-Infection: T Cell Profile, Cytokine Dynamics and Role of Exhausted Lymphocytes. Int. J. Infect. Dis. 2021, 102, 163–169. [Google Scholar] [CrossRef]
- Krause, R.; Snyman, J.; Shi-Hsia, H.; Muema, D.; Karim, F.; Ganga, Y.; Ngoepe, A.; Zungu, Y.; Gazy, I.; Bernstein, M.; et al. HIV Skews the SARS-CoV-2 B Cell Response towards an Extrafollicular Maturation Pathway. eLife 2022, 11, e79924. [Google Scholar] [CrossRef]
- Corma-Gómez, A.; Fernández-Fuertes, M.; García, E.; Fuentes-López, A.; Gómez-Ayerbe, C.; Rivero-Juárez, A.; Domínguez, C.; Santos, M.; Viñuela, L.; Palacios, R.; et al. Severe Immunosuppression Is Related to Poorer Immunogenicity to SARS-CoV-2 Vaccines among People Living with HIV. Clin. Microbiol. Infect. 2022, 28, 1492–1498. [Google Scholar] [CrossRef]
- Hassold, N.; Brichler, S.; Ouedraogo, E.; Leclerc, D.; Carroue, S.; Gater, Y.; Alloui, C.; Carbonnelle, E.; Bouchaud, O.; Mechai, F.; et al. Impaired Antibody Response to COVID-19 Vaccination in Advanced HIV Infection. AIDS Lond. Engl. 2022, 36, F1–F5. [Google Scholar] [CrossRef]
- Woodruff, M.C.; Ramonell, R.P.; Nguyen, D.C.; Cashman, K.S.; Saini, A.S.; Haddad, N.S.; Ley, A.M.; Kyu, S.; Howell, J.C.; Ozturk, T.; et al. Extrafollicular B Cell Responses Correlate with Neutralizing Antibodies and Morbidity in COVID-19. Nat. Immunol. 2020, 21, 1506–1516. [Google Scholar] [CrossRef]
- Chen, Y.; Zuiani, A.; Fischinger, S.; Mullur, J.; Atyeo, C.; Travers, M.; Lelis, F.J.N.; Pullen, K.M.; Martin, H.; Tong, P.; et al. Quick COVID-19 Healers Sustain Anti-SARS-CoV-2 Antibody Production. Cell 2020, 183, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Karim, F.; Gazy, I.; Cele, S.; Zungu, Y.; Krause, R.; Bernstein, M.; Khan, K.; Ganga, Y.; Rodel, H.; Mthabela, N.; et al. HIV Status Alters Disease Severity and Immune Cell Responses in Beta Variant SARS-CoV-2 Infection Wave. eLife 2021, 10, e67397. [Google Scholar] [CrossRef]
- Lagathu, C.; Cossarizza, A.; Béréziat, V.; Nasi, M.; Capeau, J.; Pinti, M. Basic Science and Pathogenesis of Ageing with HIV: Potential Mechanisms and Biomarkers. AIDS Lond. Engl. 2017, 31 (Suppl. 2), S105–S119. [Google Scholar] [CrossRef]
- Ho, H.; Peluso, M.J.; Margus, C.; Matias Lopes, J.P.; He, C.; Gaisa, M.M.; Osorio, G.; Aberg, J.A.; Mullen, M.P. Clinical Outcomes and Immunologic Characteristics of Coronavirus Disease 2019 in People with Human Immunodeficiency Virus. J. Infect. Dis. 2021, 223, 403–408. [Google Scholar] [CrossRef]
- Rosenthal, E.M.; Rosenberg, E.S.; Patterson, W.; Ferguson, W.P.; Gonzalez, C.; DeHovitz, J.; Udo, T.; Rajulu, D.T.; Hart-Malloy, R.; Tesoriero, J. Factors Associated with SARS-CoV-2-Related Hospital Outcomes among and between Persons Living with and without Diagnosed HIV Infection in New York State. PLoS ONE 2022, 17, e0268978. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035. [Google Scholar] [CrossRef]
- Killip, M.J.; Fodor, E.; Randall, R.E. Influenza Virus Activation of the Interferon System. Virus Res. 2015, 209, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swets, M.C.; Russell, C.D.; Harrison, E.M.; Docherty, A.B.; Lone, N.; Girvan, M.; Hardwick, H.E.; Visser, L.G.; Openshaw, P.J.M.; Groeneveld, G.H.; et al. SARS-CoV-2 Co-Infection with Influenza Viruses, Respiratory Syncytial Virus, or Adenoviruses. Lancet 2022, 399, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Stowe, J.; Tessier, E.; Zhao, H.; Guy, R.; Muller-Pebody, B.; Zambon, M.; Andrews, N.; Ramsay, M.; Lopez Bernal, J. Interactions between SARS-CoV-2 and Influenza, and the Impact of Coinfection on Disease Severity: A Test-Negative Design. Int. J. Epidemiol. 2021, 50, 1124–1133. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Chen, C.; Li, Y.; Yan, D.; Zhang, X.; Jiang, D.; Yang, S.; Li, L. Impact of Coinfection with SARS-CoV-2 and Influenza on Disease Severity: A Systematic Review and Meta-Analysis. Front. Public Health 2021, 9, 1944. [Google Scholar] [CrossRef]
- Ma, S.; Lai, X.; Chen, Z.; Tu, S.; Qin, K. Clinical Characteristics of Critically Ill Patients Co-Infected with SARS-CoV-2 and the Influenza Virus in Wuhan, China. Int. J. Infect. Dis. 2020, 96, 683–687. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, F.; Wu, K.; Wang, J.; Li, F.; Huang, S.; Lu, J.; Huang, J.; Liu, H.; Zhou, R.; et al. Clinical and Virological Impact of Single and Dual Infections with Influenza A (H1N1) and SARS-CoV-2 in Adult Inpatients. PLoS Negl. Trop. Dis. 2021, 15, e0009997. [Google Scholar] [CrossRef]
- Kinoshita, T.; Watanabe, K.; Sakurai, Y.; Nishi, K.; Yoshikawa, R.; Yasuda, J. Co-Infection of SARS-CoV-2 and Influenza Virus Causes More Severe and Prolonged Pneumonia in Hamsters. Sci. Rep. 2021, 11, 21259. [Google Scholar] [CrossRef]
- Bai, L.; Zhao, Y.; Dong, J.; Liang, S.; Guo, M.; Liu, X.; Wang, X.; Huang, Z.; Sun, X.; Zhang, Z.; et al. Coinfection with Influenza A Virus Enhances SARS-CoV-2 Infectivity. Cell Res. 2021, 31, 395–403. [Google Scholar] [CrossRef]
- Achdout, H.; Vitner, E.B.; Politi, B.; Melamed, S.; Yahalom-Ronen, Y.; Tamir, H.; Erez, N.; Avraham, R.; Weiss, S.; Cherry, L.; et al. Increased Lethality in Influenza and SARS-CoV-2 Coinfection Is Prevented by Influenza Immunity but Not SARS-CoV-2 Immunity. Nat. Commun. 2021, 12, 5819. [Google Scholar] [CrossRef]
- Fink, G.; Orlova-Fink, N.; Schindler, T.; Grisi, S.; Ferrer, A.P.S.; Daubenberger, C.; Brentani, A. Inactivated Trivalent Influenza Vaccination Is Associated with Lower Mortality among Patients with COVID-19 in Brazil. BMJ Evid.-Based Med. 2021, 26, 192–193. [Google Scholar] [CrossRef]
- Behrouzi, B.; Araujo Campoverde, M.V.; Liang, K.; Talbot, H.K.; Bogoch, I.I.; McGeer, A.; Fröbert, O.; Loeb, M.; Vardeny, O.; Solomon, S.D.; et al. Influenza Vaccination to Reduce Cardiovascular Morbidity and Mortality in Patients with COVID-19: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2020, 76, 1777–1794. [Google Scholar] [CrossRef] [PubMed]
- Fage, C.; Hénaut, M.; Carbonneau, J.; Piret, J.; Boivin, G. Influenza A(H1N1)Pdm09 Virus but Not Respiratory Syncytial Virus Interferes with SARS-CoV-2 Replication during Sequential Infections in Human Nasal Epithelial Cells. Viruses 2022, 14, 395. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Nakajima, N.; Sato, Y.; Takahashi, K.; Accola, M.; Chiba, S.; Fan, S.; Neumann, G.; Rehrauer, W.; Suzuki, T.; et al. SARS-CoV-2 Interference of Influenza Virus Replication in Syrian Hamsters. J. Infect. Dis. 2022, 225, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.C.; Lai, Y.; Barrow, K.A.; Hamerman, J.A.; Lacy-Hulbert, A.; Piliponsky, A.M.; Ziegler, S.F.; Altemeier, W.A.; Debley, J.S.; Gharib, S.A.; et al. Effects of Asthma and Human Rhinovirus A16 on the Expression of SARS-CoV-2 Entry Factors in Human Airway Epithelium. Am. J. Respir. Cell Mol. Biol. 2020, 63, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Scotta, M.C.; Kern, L.B.; Polese-Bonatto, M.; Azevedo, T.R.; Varela, F.H.; Zavaglia, G.O.; Fernandes, I.R.; de David, C.N.; Fazolo, T.; da Costa, M.S.C.; et al. Impact of Rhinovirus on Hospitalization during the COVID-19 Pandemic: A Prospective Cohort Study. J. Clin. Virol. 2022, 156, 105197. [Google Scholar] [CrossRef]
- Le Glass, E.; Hoang, V.T.; Boschi, C.; Ninove, L.; Zandotti, C.; Boutin, A.; Bremond, V.; Dubourg, G.; Ranque, S.; Lagier, J.-C.; et al. Incidence and Outcome of Coinfections with SARS-CoV-2 and Rhinovirus. Viruses 2021, 13, 2528. [Google Scholar] [CrossRef] [PubMed]
- Dee, K.; Goldfarb, D.M.; Haney, J.; Amat, J.A.R.; Herder, V.; Stewart, M.; Szemiel, A.M.; Baguelin, M.; Murcia, P.R. Human Rhinovirus Infection Blocks Severe Acute Respiratory Syndrome Coronavirus 2 Replication within the Respiratory Epithelium: Implications for COVID-19 Epidemiology. J. Infect. Dis. 2021, 224, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Vanderwall, E.R.; Barrow, K.A.; Rich, L.M.; Read, D.F.; Trapnell, C.; Okoloko, O.; Ziegler, S.F.; Hallstrand, T.S.; White, M.P.; Debley, J.S. Airway Epithelial Interferon Response to SARS-CoV-2 Is Inferior to Rhinovirus and Heterologous Rhinovirus Infection Suppresses SARS-CoV-2 Replication. Sci. Rep. 2022, 12, 6972. [Google Scholar] [CrossRef]
- Soni, S.; Namdeo Pudake, R.; Jain, U.; Chauhan, N. A Systematic Review on SARS-CoV-2-Associated Fungal Coinfections. J. Med. Virol. 2022, 94, 99–109. [Google Scholar] [CrossRef]
- Amin, A.; Vartanian, A.; Poladian, N.; Voloshko, A.; Yegiazaryan, A.; Al-Kassir, A.L.; Venketaraman, V. Root Causes of Fungal Coinfections in COVID-19 Infected Patients. Infect. Dis. Rep. 2021, 13, 1018–1035. [Google Scholar] [CrossRef]
- Salmanton-García, J.; Sprute, R.; Stemler, J.; Bartoletti, M.; Dupont, D.; Valerio, M.; Garcia-Vidal, C.; Falces-Romero, I.; Machado, M.; de la Villa, S.; et al. COVID-19-Associated Pulmonary Aspergillosis, March–August 2020. Emerg. Infect. Dis. 2021, 27, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-C.; Yu, W.-L. COVID-19 Associated with Pulmonary Aspergillosis: A Literature Review. J. Microbiol. Immunol. Infect. 2021, 54, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.-P.; Tang, Y.-M.; Song, H.; Xu, W.-Q.; Yang, S.-L.; Xu, X.-J. Efficiency of Interleukin 6 and Interferon Gamma in the Differentiation of Invasive Pulmonary Aspergillosis and Pneumocystis Pneumonia in Pediatric Oncology Patients. Int. J. Infect. Dis. 2016, 48, 73–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camargo, J.F.; Bhimji, A.; Kumar, D.; Kaul, R.; Pavan, R.; Schuh, A.; Seftel, M.; Lipton, J.H.; Gupta, V.; Humar, A.; et al. Impaired T Cell Responsiveness to Interleukin-6 in Hematological Patients with Invasive Aspergillosis. PLoS ONE 2015, 10, e0123171. [Google Scholar] [CrossRef]
- Clemons, K.V.; Grunig, G.; Sobel, R.A.; Mirels, L.F.; Rennick, D.M.; Stevens, D.A. Role of IL-10 in Invasive Aspergillosis: Increased Resistance of IL-10 Gene Knockout Mice to Lethal Systemic Aspergillosis. Clin. Exp. Immunol. 2000, 122, 186–191. [Google Scholar] [CrossRef]
- Del Sero, G.; Mencacci, A.; Cenci, E.; Fè d’Ostiani, C.; Montagnoli, C.; Bacci, A.; Mosci, P.; Kopf, M.; Romani, L. Antifungal Type 1 Responses Are Upregulated in IL-10-Deficient Mice. Microbes Infect. 1999, 1, 1169–1180. [Google Scholar] [CrossRef]
- Segrelles-Calvo, G.; de S Araújo, G.R.; Llopis-Pastor, E.; Carrillo, J.; Hernández-Hernández, M.; Rey, L.; Melean, N.R.; Escribano, I.; Antón, E.; Zamarro, C.; et al. Candida Spp. Co-Infection in COVID-19 Patients with Severe Pneumonia: Prevalence Study and Associated Risk Factors. Respir. Med. 2021, 188, 106619. [Google Scholar] [CrossRef]
- Castro, M.; Bjoraker, J.A.; Rohrbach, M.S.; Limper, A.H. Candida Albicans Induces the Release of Inflammatory Mediators from Human Peripheral Blood Monocytes. Inflammation 1996, 20, 107–122. [Google Scholar] [CrossRef]
- Steinshamn, S.; Waage, A. Tumor Necrosis Factor and Interleukin-6 in Candida Albicans Infection in Normal and Granulocytopenic Mice. Infect. Immun. 1992, 60, 4003–4008. [Google Scholar] [CrossRef]
- Campbell, L.; Hepworth, M.R.; Whittingham-Dowd, J.; Thompson, S.; Bancroft, A.J.; Hayes, K.S.; Shaw, T.N.; Dickey, B.F.; Flamar, A.-L.; Artis, D.; et al. ILC2s Mediate Systemic Innate Protection by Priming Mucus Production at Distal Mucosal Sites. J. Exp. Med. 2019, 216, 2714–2723. [Google Scholar] [CrossRef] [Green Version]
- Wolday, D.; Gebrecherkos, T.; Arefaine, Z.G.; Kiros, Y.K.; Gebreegzabher, A.; Tasew, G.; Abdulkader, M.; Abraha, H.E.; Desta, A.A.; Hailu, A.; et al. Effect of Co-Infection with Intestinal Parasites on COVID-19 Severity: A Prospective Observational Cohort Study. EClinicalMedicine 2021, 39, 101054. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Mu, Y.; McManus, D.P. The Fight Against Severe COVID-19: Can Parasitic Worms Contribute? Front. Immunol. 2022, 13, 849465. [Google Scholar] [CrossRef] [PubMed]
- Ademe, M.; Girma, F. The Influence of Helminth Immune Regulation on COVID-19 Clinical Outcomes: Is It Beneficial or Detrimental? Infect. Drug Resist. 2021, 14, 4421–4426. [Google Scholar] [CrossRef]
- Wolday, D.; Tasew, G.; Amogne, W.; Urban, B.; Schallig, H.D.; Harris, V.; Rinke de Wit, T.F. Interrogating the Impact of Intestinal Parasite-Microbiome on Pathogenesis of COVID-19 in Sub-Saharan Africa. Front. Microbiol. 2021, 12, 614522. [Google Scholar] [CrossRef] [PubMed]
- Paniz-Mondolfi, A.E.; Ramírez, J.D.; Delgado-Noguera, L.A.; Rodriguez-Morales, A.J.; Sordillo, E.M. COVID-19 and Helminth Infection: Beyond the Th1/Th2 Paradigm. PLoS Negl. Trop. Dis. 2021, 15, e0009402. [Google Scholar] [CrossRef]
- Hussein, R.; Guedes, M.; Ibraheim, N.; Ali, M.M.; El-Tahir, A.; Allam, N.; Abuakar, H.; Pecoits-Filho, R.; Kotanko, P. Impact of COVID-19 and Malaria Coinfection on Clinical Outcomes: A Retrospective Cohort Study. Clin. Microbiol. Infect. 2022, 28, 1152.e1–1152.e6. [Google Scholar] [CrossRef]
- Herrmann, M.; Schulte, S.; Wildner, N.H.; Wittner, M.; Brehm, T.T.; Ramharter, M.; Woost, R.; Lohse, A.W.; Jacobs, T.; Schulze zur Wiesch, J. Analysis of Co-Inhibitory Receptor Expression in COVID-19 Infection Compared to Acute Plasmodium Falciparum Malaria: LAG-3 and TIM-3 Correlate with T Cell Activation and Course of Disease. Front. Immunol. 2020, 11, 1870. [Google Scholar] [CrossRef]
- Wildner, N.H.; Ahmadi, P.; Schulte, S.; Brauneck, F.; Kohsar, M.; Lütgehetmann, M.; Beisel, C.; Addo, M.M.; Haag, F.; Schulze Zur Wiesch, J. B Cell Analysis in SARS-CoV-2 versus Malaria: Increased Frequencies of Plasmablasts and Atypical Memory B Cells in COVID-19. J. Leukoc. Biol. 2021, 109, 77–90. [Google Scholar] [CrossRef]
- Mahajan, N.N.; Gajbhiye, R.K.; Bahirat, S.; Lokhande, P.D.; Mathe, A.; Rathi, S.; Warty, N.; Mahajan, K.N.; Srivastava, V.; Kuppusamy, P.; et al. Co-Infection of Malaria and Early Clearance of SARS-CoV-2 in Healthcare Workers. J. Med. Virol. 2021, 93, 2431–2438. [Google Scholar] [CrossRef]
- Molina, I.; Marcolino, M.S.; Pires, M.C.; Ramos, L.E.F.; Silva, R.T.; Guimarães-Júnior, M.H.; de Oliveira, I.J.R.; de Carvalho, R.L.R.; Nunes, A.G.S.; de Barros, A.L.R.M.; et al. Chagas Disease and SARS-CoV-2 Coinfection Does Not Lead to Worse in-Hospital Outcomes. Sci. Rep. 2021, 11, 20289. [Google Scholar] [CrossRef]
- Fernandez, M.L.; Benchetrit, A.; Astudillo, O.G.; Garay, A.M.; De Vedia, L.; Garcia Bournissen, F.; Lloveras, S.C.; Orduna, T.A.; Gonzalez, G.D. COVID-19 and Chagas Disease in Buenos Aires, Argentina. Front. Trop. Dis. 2022, 2, 779428. [Google Scholar] [CrossRef]
- Golda, A.; Malek, N.; Dudek, B.; Zeglen, S.; Wojarski, J.; Ochman, M.; Kucewicz, E.; Zembala, M.; Potempa, J.; Pyrc, K. 2011 Infection with Human Coronavirus NL63 Enhances Streptococcal Adherence to Epithelial Cells. J. Gen. Virol. 2011, 92, 1358–1368. [Google Scholar] [CrossRef] [PubMed]
- Ramphal, R.; Small, P.M.; Shands, J.W.; Fischlschweiger, W.; Small, P.A. Adherence of Pseudomonas Aeruginosa to Tracheal Cells Injured by Influenza Infection or by Endotracheal Intubation. Infect. Immun. 1980, 27, 614–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowe, H.M.; Meliopoulos, V.A.; Iverson, A.; Bomme, P.; Schultz-Cherry, S.; Rosch, J.W. Direct Interactions with Influenza Promote Bacterial Adherence during Respiratory Infections. Nat. Microbiol. 2019, 4, 1328–1336. [Google Scholar] [CrossRef] [PubMed]
- Robinot, R.; Hubert, M.; de Melo, G.D.; Lazarini, F.; Bruel, T.; Smith, N.; Levallois, S.; Larrous, F.; Fernandes, J.; Gellenoncourt, S.; et al. SARS-CoV-2 Infection Induces the Dedifferentiation of Multiciliated Cells and Impairs Mucociliary Clearance. Nat. Commun. 2021, 12, 4354. [Google Scholar] [CrossRef] [PubMed]
- Avadhanula, V.; Rodriguez, C.A.; De Vincenzo, J.P.; Wang, Y.; Webby, R.J.; Ulett, G.C.; Adderson, E.E. Respiratory Viruses Augment the Adhesion of Bacterial Pathogens to Respiratory Epithelium in a Viral Species- and Cell Type-Dependent Manner. J. Virol. 2006, 80, 1629–1636. [Google Scholar] [CrossRef] [Green Version]
- Larriva, M.A.-D.; Martín-DeLeon, R.; Royo, B.U.; Fernández-Navamuel, I.; Velando, A.G.; García, L.N.; Clemente, C.C.; García, F.A.; Codern, A.R.; Fernández-Arias, C.; et al. The Role of Bronchoscopy in Patients with SARS-CoV-2 Pneumonia. ERJ Open Res. 2021, 7, 00165. [Google Scholar] [CrossRef]
- Meyerholz, D.K.; Reznikov, L.R. Influence of SARS-CoV-2 on Airway Mucus Production: A Review and Proposed Model. Vet. Pathol. 2022, 59, 578–585. [Google Scholar] [CrossRef]
- Østergaard, L. SARS CoV-2 Related Microvascular Damage and Symptoms during and after COVID-19: Consequences of Capillary Transit-Time Changes, Tissue Hypoxia and Inflammation. Physiol. Rep. 2021, 9, e14726. [Google Scholar] [CrossRef]
- Noel-Savina, E.; Viatgé, T.; Faviez, G.; Lepage, B.; Mhanna, L.T.; Pontier, S.; Dupuis, M.; Collot, S.; Thomas, P.; Idoate Lacasia, J.; et al. Severe SARS-CoV-2 Pneumonia: Clinical, Functional and Imaging Outcomes at 4 Months. Respir. Med. Res. 2021, 80, 100822. [Google Scholar] [CrossRef]
- Barbeta, E.; Motos, A.; Torres, A.; Ceccato, A.; Ferrer, M.; Cilloniz, C.; Bueno, L.; Badia, J.R.; Castro, P.; Ferrando, C.; et al. SARS-CoV-2–Induced Acute Respiratory Distress Syndrome: Pulmonary Mechanics and Gas-Exchange Abnormalities. Ann. Am. Thorac. Soc. 2020, 17, 1164–1168. [Google Scholar] [CrossRef]
- Ackermann, M.; Anders, H.-J.; Bilyy, R.; Bowlin, G.L.; Daniel, C.; De Lorenzo, R.; Egeblad, M.; Henneck, T.; Hidalgo, A.; Hoffmann, M.; et al. Patients with COVID-19: In the Dark-NETs of Neutrophils. Cell Death Differ. 2021, 28, 3125–3139. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Mogensen, T.H. Innate Immunological Pathways in COVID-19 Pathogenesis. Sci. Immunol. 2022, 7, eabm5505. [Google Scholar] [CrossRef] [PubMed]
- Feldman, C.; Anderson, R. The Role of Co-Infections and Secondary Infections in Patients with COVID-19. Pneumonia 2021, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liu, W.; Jiang, M.; Huang, P.; Xiang, Z.; Deng, D.; Chen, P.; Xie, L. Clinical Characteristics of COVID-19 Patients with Clinically Diagnosed Bacterial Co-Infection: A Multi-Center Study. PLoS ONE 2021, 16, e0249668. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, I.; Chung, M.; Angel, L.; Tsay, J.-C.J.; Wu, B.G.; Yeung, S.T.; Krolikowski, K.; Li, Y.; Duerr, R.; Schluger, R.; et al. Microbial Signatures in the Lower Airways of Mechanically Ventilated COVID-19 Patients Associated with Poor Clinical Outcome. Nat. Microbiol. 2021, 6, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Cele, S.; Karim, F.; Lustig, G.; San, J.E.; Hermanus, T.; Tegally, H.; Snyman, J.; Moyo-Gwete, T.; Wilkinson, E.; Bernstein, M.; et al. SARS-CoV-2 Prolonged Infection during Advanced HIV Disease Evolves Extensive Immune Escape. Cell Host Microbe 2022, 30, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Song, J.; Dai, L.; Post, S.R.; Qin, Z. SARS-CoV-2 Infection and Lytic Reactivation of Herpesviruses: A Potential Threat in the Postpandemic Era? J. Med. Virol. 2022, 94, 5103–5111. [Google Scholar] [CrossRef] [PubMed]
- Pathak, L.; Gayan, S.; Pal, B.; Talukdar, J.; Bhuyan, S.; Sandhya, S.; Yeger, H.; Baishya, D.; Das, B. Coronavirus Activates an Altruistic Stem Cell-Mediated Defense Mechanism That Reactivates Dormant Tuberculosis: Implications in Coronavirus Disease 2019 Pandemic. Am. J. Pathol. 2021, 191, 1255–1268. [Google Scholar] [CrossRef]
- Pakzad, R.; Malekifar, P.; Shateri, Z.; Zandi, M.; Akhavan Rezayat, S.; Soleymani, M.; Karimi, M.R.; Ahmadi, S.E.; Shahbahrami, R.; Pakzad, I.; et al. Worldwide Prevalence of Microbial Agents’ Coinfection among COVID-19 Patients: A Comprehensive Updated Systematic Review and Meta-Analysis. J. Clin. Lab. Anal. 2022, 36, e24151. [Google Scholar] [CrossRef]
- Alhumaid, S.; Al Mutair, A.; Al Alawi, Z.; Alshawi, A.M.; Alomran, S.A.; Almuhanna, M.S.; Almuslim, A.A.; Bu Shafia, A.H.; Alotaibi, A.M.; Ahmed, G.Y.; et al. Coinfections with Bacteria, Fungi, and Respiratory Viruses in Patients with SARS-CoV-2: A Systematic Review and Meta-Analysis. Pathogens 2021, 10, 809. [Google Scholar] [CrossRef]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden, D.R.; Soucy, J.-P.R.; Daneman, N. Bacterial Co-Infection and Secondary Infection in Patients with COVID-19: A Living Rapid Review and Meta-Analysis. Clin. Microbiol. Infect. 2020, 26, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Kollef, M.H.; Timsit, J.-F. Bacterial and Fungal Superinfections in Critically Ill Patients with COVID-19. Intensive Care Med. 2020, 46, 2071–2074. [Google Scholar] [CrossRef] [PubMed]
- Malekifar, P.; Pakzad, R.; Shahbahrami, R.; Zandi, M.; Jafarpour, A.; Rezayat, S.A.; Akbarpour, S.; Shabestari, A.N.; Pakzad, I.; Hesari, E.; et al. Viral Coinfection among COVID-19 Patient Groups: An Update Systematic Review and Meta-Analysis. BioMed Res. Int. 2021, 2021, 5313832. [Google Scholar] [CrossRef] [PubMed]
- Krumbein, H.; Kümmel, L.S.; Fragkou, P.C.; Thölken, C.; Hünerbein, B.L.; Reiter, R.; Papathanasiou, K.A.; Renz, H.; Skevaki, C. Respiratory Viral Co-Infections in Patients with COVID-19 and Associated Outcomes: A Systematic Review and Meta-Analysis. Rev. Med. Virol. 2022, e2365. [Google Scholar] [CrossRef]
- Singh, V.; Upadhyay, P.; Reddy, J.; Granger, J. SARS-CoV-2 Respiratory Co-Infections: Incidence of Viral and Bacterial Co-Pathogens. Int. J. Infect. Dis. 2021, 105, 617–620. [Google Scholar] [CrossRef]
- Hoque, M.N.; Akter, S.; Mishu, I.D.; Islam, M.R.; Rahman, M.S.; Akhter, M.; Islam, I.; Hasan, M.M.; Rahaman, M.M.; Sultana, M.; et al. Microbial Co-Infections in COVID-19: Associated Microbiota and Underlying Mechanisms of Pathogenesis. Microb. Pathog. 2021, 156, 104941. [Google Scholar] [CrossRef]
- Goldberg, E.M.; Hasegawa, K.; Lawrence, A.; Kline, J.A.; Camargo, C.A. Viral Coinfection Is Associated with Improved Outcomes in Emergency Department Patients with SARS-CoV-2. West. J. Emerg. Med. 2021, 22, 1262–1269. [Google Scholar] [CrossRef]
- Chekuri, S.; Szymczak, W.A.; Goldstein, D.Y.; Nori, P.; Marrero Rolon, R.; Spund, B.; Singh-Tan, S.; Mohrmann, L.; Assa, A.; Southern, W.N.; et al. SARS-CoV-2 Coinfection with Additional Respiratory Virus Does Not Predict Severe Disease: A Retrospective Cohort Study. J. Antimicrob. Chemother. 2021, 76, iii12–iii19. [Google Scholar] [CrossRef]
- Kumar, N.; Sharma, S.; Barua, S.; Tripathi, B.N.; Rouse, B.T. Virological and Immunological Outcomes of Coinfections. Clin. Microbiol. Rev. 2018, 31, e00111-17. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Wang, Q.; Mei, H.; Zheng, H.; Liang, G.; She, X.; Liu, W. Fungal Co-Infection in COVID-19 Patients: Evidence from a Systematic Review and Meta-Analysis. Aging 2021, 13, 7745–7757. [Google Scholar] [CrossRef]
- Sreenath, K.; Batra, P.; Vinayaraj, E.V.; Bhatia, R.; SaiKiran, K.; Singh, V.; Singh, S.; Verma, N.; Singh, U.B.; Mohan, A.; et al. Coinfections with Other Respiratory Pathogens among Patients with COVID-19. Microbiol. Spectr. 2021, 9, e00163-21. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Chua, J.V.; Baddley, J.W. Coronavirus Disease 2019-Associated Mucormycosis: Risk Factors and Mechanisms of Disease. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2022, 74, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Muthu, V.; Rudramurthy, S.M.; Chakrabarti, A.; Agarwal, R. Epidemiology and Pathophysiology of COVID-19-Associated Mucormycosis: India Versus the Rest of the World. Mycopathologia 2021, 186, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamed, E.F.; Ibrahim, M.N.; Mostafa, N.E.; Moawad, H.S.F.; Elgammal, N.E.; Darwiesh, E.M.; El-rafey, D.S.; ElBadawy, N.E.; Al-Khoufi, E.A.; Hindawi, S.I. Role of Interferon Gamma in SARS-CoV-2-Positive Patients with Parasitic Infections. Gut Pathog. 2021, 13, 29. [Google Scholar] [CrossRef]
- Wilairatana, P.; Masangkay, F.R.; Kotepui, K.U.; Milanez, G.D.J.; Kotepui, M. Prevalence and Characteristics of Malaria among COVID-19 Individuals: A Systematic Review, Meta-Analysis, and Analysis of Case Reports. PLoS Negl. Trop. Dis. 2021, 15, e0009766. [Google Scholar] [CrossRef]
- Leng, S.; Chen, X.; Mao, G. Frailty Syndrome: An Overview. Clin. Interv. Aging 2014, 9, 433. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. WHO Clinical Consortium on Healthy Ageing (Topic Focus: Frailty and Intrinsic Capacity); World Health Organization: Geneva, Switzerland, 2016; p. 36. [Google Scholar]
- Morley, J.E.; Vellas, B.; Kan, G.A.V.; Anker, S.D.; Bauer, J.M.; Bernabei, R.; Cesari, M.; Chumlea, W.C.; Doehner, W.; Evans, J.; et al. Frailty Consensus: A Call to Action. J. Am. Med. Dir. Assoc. 2013, 14, 392–397. [Google Scholar] [CrossRef] [Green Version]
- O’Caoimh, R.; Sezgin, D.; O’Donovan, M.R.; Molloy, D.W.; Clegg, A.; Rockwood, K.; Liew, A. Prevalence of Frailty in 62 Countries across the World: A Systematic Review and Meta-Analysis of Population-Level Studies. Age Ageing 2021, 50, 96–104. [Google Scholar] [CrossRef]
- Kojima, G. Prevalence of Frailty in Nursing Homes: A Systematic Review and Meta-Analysis. J. Am. Med. Dir. Assoc. 2015, 16, 940–945. [Google Scholar] [CrossRef]
- Collard, R.M.; Boter, H.; Schoevers, R.A.; Oude Voshaar, R.C. Prevalence of Frailty in Community-Dwelling Older Persons: A Systematic Review. J. Am. Geriatr. Soc. 2012, 60, 1487–1492. [Google Scholar] [CrossRef]
- Rockwood, K.; Song, X.; MacKnight, C.; Bergman, H.; Hogan, D.B.; McDowell, I.; Mitnitski, A. A Global Clinical Measure of Fitness and Frailty in Elderly People. CMAJ 2005, 173, 489–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewitt, J.; Carter, B.; Vilches-Moraga, A.; Quinn, T.J.; Braude, P.; Verduri, A.; Pearce, L.; Stechman, M.; Short, R.; Price, A.; et al. The Effect of Frailty on Survival in Patients with COVID-19 (COPE): A Multicentre, European, Observational Cohort Study. Lancet Public Health 2020, 5, e444–e451. [Google Scholar] [CrossRef] [PubMed]
- Sablerolles, R.S.G.; Lafeber, M.; van Kempen, J.A.L.; van de Loo, B.P.A.; Boersma, E.; Rietdijk, W.J.R.; Polinder-Bos, H.A.; Mooijaart, S.P.; van der Kuy, H.; Versmissen, J.; et al. Association between Clinical Frailty Scale Score and Hospital Mortality in Adult Patients with COVID-19 (COMET): An International, Multicentre, Retrospective, Observational Cohort Study. Lancet Healthy Longev. 2021, 2, e163–e170. [Google Scholar] [CrossRef] [PubMed]
- Kundi, H.; Çetin, E.H.Ö.; Canpolat, U.; Aras, S.; Celik, O.; Ata, N.; Birinci, S.; Çay, S.; Özeke, Ö.; Tanboğa, I.H.; et al. The Role of Frailty on Adverse Outcomes among Older Patients with COVID-19. J. Infect. 2020, 81, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Izurieta, H.S.; Graham, D.J.; Jiao, Y.; Hu, M.; Lu, Y.; Wu, Y.; Chillarige, Y.; Wernecke, M.; Menis, M.; Pratt, D.; et al. Natural History of Coronavirus Disease 2019: Risk Factors for Hospitalizations and Deaths among >26 Million US Medicare Beneficiaries. J. Infect. Dis. 2021, 223, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Kastora, S.; Kounidas, G.; Perrott, S.; Carter, B.; Hewitt, J.; Myint, P.K. Clinical Frailty Scale as a Point of Care Prognostic Indicator of Mortality in COVID-19: A Systematic Review and Meta-Analysis. EClinicalMedicine 2021, 36, 100896. [Google Scholar] [CrossRef] [PubMed]
- Rottler, M.; Ocskay, K.; Sipos, Z.; Görbe, A.; Virág, M.; Hegyi, P.; Molnár, T.; Erőss, B.; Leiner, T.; Molnár, Z. Clinical Frailty Scale (CFS) Indicated Frailty Is Associated with Increased in-Hospital and 30-Day Mortality in COVID-19 Patients: A Systematic Review and Meta-Analysis. Ann. Intensive Care 2022, 12, 17. [Google Scholar] [CrossRef]
- Blomaard, L.C.; van der Linden, C.M.J.; van der Bol, J.M.; Jansen, S.W.M.; Polinder-Bos, H.A.; Willems, H.C.; Festen, J.; Barten, D.G.; Borgers, A.J.; Bos, J.C.; et al. Frailty Is Associated with In-Hospital Mortality in Older Hospitalised COVID-19 Patients in the Netherlands: The COVID-OLD Study. Age Ageing 2021, 50, 631–640. [Google Scholar] [CrossRef]
- Subramaniam, A.; Anstey, C.; Curtis, J.R.; Ashwin, S.; Ponnapa Reddy, M.; Aliberti, M.J.R.; Avelino-Silva, T.J.; Welch, C.; Koduri, G.; Prowle, J.R.; et al. Characteristics and Outcomes of Patients with Frailty Admitted to ICU with Coronavirus Disease 2019: An Individual Patient Data Meta-Analysis. Crit. Care Explor. 2022, 4, e0616. [Google Scholar] [CrossRef]
- Taniguchi, L.U.; Avelino-Silva, T.J.; Dias, M.B.; Jacob-Filho, W.; Aliberti, M.J.R.; CO-FRAIL Study Group; EPICCoV Study Group; COVID HCFMUSP Study Group. Association of Frailty, Organ Support, and Long-Term Survival in Critically Ill Patients with COVID-19. Crit. Care Explor. 2022, 4, e0712. [Google Scholar] [CrossRef]
- Pranata, R.; Henrina, J.; Lim, M.A.; Lawrensia, S.; Yonas, E.; Vania, R.; Huang, I.; Lukito, A.A.; Suastika, K.; Kuswardhani, R.A.T.; et al. Clinical Frailty Scale and Mortality in COVID-19: A Systematic Review and Dose-Response Meta-Analysis. Arch. Gerontol. Geriatr. 2021, 93, 104324. [Google Scholar] [CrossRef] [PubMed]
- Kojima, G. Frailty Defined by FRAIL Scale as a Predictor of Mortality: A Systematic Review and Meta-Analysis. J. Am. Med. Dir. Assoc. 2018, 19, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Kojima, G.; Iliffe, S.; Walters, K. Frailty Index as a Predictor of Mortality: A Systematic Review and Meta-Analysis. Age Ageing 2018, 47, 193–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanhella, K.J.; Fernandez-Patron, C. Biomarkers of Ageing and Frailty May Predict COVID-19 Severity. Ageing Res. Rev. 2022, 73, 101513. [Google Scholar] [CrossRef]
- Hussien, H.; Nastasa, A.; Apetrii, M.; Nistor, I.; Petrovic, M.; Covic, A. Different Aspects of Frailty and COVID-19: Points to Consider in the Current Pandemic and Future Ones. BMC Geriatr. 2021, 21, 389. [Google Scholar] [CrossRef]
- She, Q.; Chen, B.; Liu, W.; Li, M.; Zhao, W.; Wu, J. Frailty Pathogenesis, Assessment, and Management in Older Adults with COVID-19. Front. Med. 2021, 8, 694367. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Haiminen, N.; Utro, F.; Seabolt, E.; Parida, L. Functional Profiling of COVID-19 Respiratory Tract Microbiomes. Sci. Rep. 2021, 11, 6433. [Google Scholar] [CrossRef]
- Servin, A.L. Antagonistic Activities of Lactobacilli and Bifidobacteria against Microbial Pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [Green Version]
- Bodera, P.; Chcialowski, A. Immunomodulatory Effect of Probiotic Bacteria. Recent Pat. Inflamm. Allergy Drug Discov. 2009, 3, 58–64. [Google Scholar] [CrossRef]
- Vallianou, N.; Stratigou, T.; Christodoulatos, G.S.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Obesity and Obesity-Associated Metabolic Disorders: Current Evidence and Perspectives. Curr. Obes. Rep. 2019, 8, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, H.S.; Bilen, Ö. Oral Booster Probiotic Bifidobacteria in SARS-COV-2 Patients. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211059676. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Cheng, F.; Xu, Q.; Su, Y.; Cai, X.; Zeng, F.; Zhang, Y. The Role of Probiotics in Coronavirus Disease-19 Infection in Wuhan: A Retrospective Study of 311 Severe Patients. Int. Immunopharmacol. 2021, 95, 107531. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.; Ashoush, O.A.; Hegazy, M.T.; Wahba, M.; Lithy, R.M.; Abdel-Hamid, H.M.; Elshafy, S.A.A.; Abdelfatah, D.; Ibrahim, M.H.E.-D.; Abdelghani, A. Beyond Probiotic Legend: ESSAP Gut Microbiota Health Score to Delineate SARS-COV-2 Infection Severity. Br. J. Nutr. 2022, 127, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Llor, C.; Ouchi, D.; Giner-Soriano, M.; García-Sangenís, A.; Bjerrum, L.; Morros, R. Correlation between Previous Antibiotic Exposure and COVID-19 Severity. A Population-Based Cohort Study. Antibiotics 2021, 10, 1364. [Google Scholar] [CrossRef]
- Yin, X.; Xu, X.; Li, H.; Jiang, N.; Wang, J.; Lu, Z.; Xiong, N.; Gong, Y. Evaluation of Early Antibiotic Use in Patients with Non-Severe COVID-19 without Bacterial Infection. Int. J. Antimicrob. Agents 2022, 59, 106462. [Google Scholar] [CrossRef]
- Man, W.H.; de Steenhuijsen Piters, W.A.A.; Bogaert, D. The Microbiota of the Respiratory Tract: Gatekeeper to Respiratory Health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human Nutrition, the Gut Microbiome and the Immune System. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef]
- Iebba, V.; Zanotta, N.; Campisciano, G.; Zerbato, V.; Di Bella, S.; Cason, C.; Luzzati, R.; Confalonieri, M.; Palamara, A.T.; Comar, M. Profiling of Oral Microbiota and Cytokines in COVID-19 Patients. Front. Microbiol. 2021, 12, 1603. [Google Scholar] [CrossRef]
- Gupta, A.; Bhanushali, S.; Sanap, A.; Shekatkar, M.; Kharat, A.; Raut, C.; Bhonde, R.; Shouche, Y.; Kheur, S.; Sharma, A. Oral Dysbiosis and Its Linkage with SARS-CoV-2 Infection. Microbiol. Res. 2022, 261, 127055. [Google Scholar] [CrossRef]
- Tchoupou Saha, O.L.F.; Dubourg, G.; Yacouba, A.; Bossi, V.; Raoult, D.; Lagier, J.-C. Profile of the Nasopharyngeal Microbiota Affecting the Clinical Course in COVID-19 Patients. Front. Microbiol. 2022, 13, 871627. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Jia, Z.; Shi, J.; Wang, W.; He, K. The Active Lung Microbiota Landscape of COVID-19 Patients through the Metatranscriptome Data Analysis. BioImpacts 2021, 12, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; Viciani, E.; Bartoletti, M.; Lewis, R.E.; Tonetti, T.; Lombardo, D.; Castagnetti, A.; Bovo, F.; Horna, C.S.; Ranieri, M.; et al. The Lower Respiratory Tract Microbiome of Critically Ill Patients with COVID-19. Sci. Rep. 2021, 11, 10103. [Google Scholar] [CrossRef] [PubMed]
- Hurst, J.H.; McCumber, A.W.; Aquino, J.N.; Rodriguez, J.; Heston, S.M.; Lugo, D.J.; Rotta, A.T.; Turner, N.A.; Pfeiffer, T.S.; Gurley, T.C.; et al. Age-Related Changes in the Nasopharyngeal Microbiome Are Associated with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection and Symptoms among Children, Adolescents, and Young Adults. Clin. Infect. Dis. 2022, 75, e928–e937. [Google Scholar] [CrossRef] [PubMed]
- Soffritti, I.; D’Accolti, M.; Fabbri, C.; Passaro, A.; Manfredini, R.; Zuliani, G.; Libanore, M.; Franchi, M.; Contini, C.; Caselli, E. Oral Microbiome Dysbiosis Is Associated with Symptoms Severity and Local Immune/Inflammatory Response in COVID-19 Patients: A Cross-Sectional Study. Front Microbiol 2021, 12, 687513. [Google Scholar] [CrossRef]
- Hoque, M.N.; Sarkar, M.M.H.; Rahman, M.S.; Akter, S.; Banu, T.A.; Goswami, B.; Jahan, I.; Hossain, M.S.; Shamsuzzaman, A.K.M.; Nafisa, T.; et al. SARS-CoV-2 Infection Reduces Human Nasopharyngeal Commensal Microbiome with Inclusion of Pathobionts. Sci. Rep. 2021, 11, 24042. [Google Scholar] [CrossRef]
- Ren, L.; Wang, Y.; Zhong, J.; Li, X.; Xiao, Y.; Li, J.; Yang, J.; Fan, G.; Guo, L.; Shen, Z.; et al. Dynamics of the Upper Respiratory Tract Microbiota and Its Association with Mortality in COVID-19. Am. J. Respir. Crit. Care Med. 2021, 204, 1379–1390. [Google Scholar] [CrossRef]
- Smith, N.; Goncalves, P.; Charbit, B.; Grzelak, L.; Beretta, M.; Planchais, C.; Bruel, T.; Rouilly, V.; Bondet, V.; Hadjadj, J.; et al. Distinct Systemic and Mucosal Immune Responses during Acute SARS-CoV-2 Infection. Nat. Immunol. 2021, 22, 1428–1439. [Google Scholar] [CrossRef]
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut through Type I Interferons. PLOS Pathog. 2016, 12, e1005572. [Google Scholar] [CrossRef] [Green Version]
- Steed, A.L.; Christophi, G.P.; Kaiko, G.E.; Sun, L.; Goodwin, V.M.; Jain, U.; Esaulova, E.; Artyomov, M.N.; Morales, D.J.; Holtzman, M.J.; et al. The Microbial Metabolite Desaminotyrosine Protects from Influenza through Type I Interferon. Science 2017, 357, 498–502. [Google Scholar] [CrossRef] [Green Version]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota Regulates Immune Defense against Respiratory Tract Influenza A Virus Infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.-Y.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.-C.; et al. Gut Microbiota Dynamics in a Prospective Cohort of Patients with Post-Acute COVID-19 Syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Xiang, B.-J.; Jiang, M.; Sun, M.; Dai, C. The Prevalence of Gastrointestinal Symptoms, Abnormal Liver Function, Digestive System Disease and Liver Disease in COVID-19 Infection. J. Clin. Gastroenterol. 2021, 55, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Zarifian, A.; Zamiri Bidary, M.; Arekhi, S.; Rafiee, M.; Gholamalizadeh, H.; Amiriani, A.; Ghaderi, M.S.; Khadem-Rezaiyan, M.; Amini, M.; Ganji, A. Gastrointestinal and Hepatic Abnormalities in Patients with Confirmed COVID-19: A Systematic Review and Meta-Analysis. J. Med. Virol. 2021, 93, 336–350. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, F.; Fahriani, M.; Mamada, S.S.; Frediansyah, A.; Abubakar, A.; Maghfirah, D.; Fajar, J.K.; Maliga, H.A.; Ilmawan, M.; Emran, T.B.; et al. Global Prevalence of Prolonged Gastrointestinal Symptoms in COVID-19 Survivors and Potential Pathogenesis: A Systematic Review and Meta-Analysis. F1000Research 2021, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Chhibber-Goel, J.; Gopinathan, S.; Sharma, A. Interplay between Severities of COVID-19 and the Gut Microbiome: Implications of Bacterial Co-Infections? Gut Pathog. 2021, 13, 14. [Google Scholar] [CrossRef]
- Zuo, T.; Zhan, H.; Zhang, F.; Liu, Q.; Tso, E.Y.K.; Lui, G.C.Y.; Chen, N.; Li, A.; Lu, W.; Chan, F.K.L.; et al. Alterations in Fecal Fungal Microbiome of Patients with COVID-19 During Time of Hospitalization until Discharge. Gastroenterology 2020, 159, 1302–1310. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, W.; Guo, M.; Xiao, C.; Fu, Z.; Yu, S.; Jiang, L.; Wang, S.; Ling, Y.; Liu, F.; et al. Integrated Analysis of Gut Microbiome and Host Immune Responses in COVID-19. Front. Med. 2022, 16, 263–275. [Google Scholar] [CrossRef]
- Ventero, M.P.; Cuadrat, R.R.C.; Vidal, I.; Andrade, B.G.N.; Molina-Pardines, C.; Haro-Moreno, J.M.; Coutinho, F.H.; Merino, E.; Regitano, L.C.A.; Silveira, C.B.; et al. Nasopharyngeal Microbial Communities of Patients Infected with SARS-CoV-2 That Developed COVID-19. Front. Microbiol. 2021, 12, 560. [Google Scholar] [CrossRef]
- Albrich, W.C.; Ghosh, T.S.; Ahearn-Ford, S.; Mikaeloff, F.; Lunjani, N.; Forde, B.; Suh, N.; Kleger, G.-R.; Pietsch, U.; Frischknecht, M.; et al. A High-Risk Gut Microbiota Configuration Associates with Fatal Hyperinflammatory Immune and Metabolic Responses to SARS-CoV-2. Gut Microbes 2022, 14, 2073131. [Google Scholar] [CrossRef]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.-Y.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.; Chan, P.K.; et al. Depicting SARS-CoV-2 Faecal Viral Activity in Association with Gut Microbiota Composition in Patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wan, Y.; Zuo, T.; Yeoh, Y.K.; Liu, Q.; Zhang, L.; Zhan, H.; Lu, W.; Xu, W.; Lui, G.C.Y.; et al. Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients with COVID-19. Gastroenterology 2022, 162, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Song, Z.-G.; Liu, C.; Tan, S.; Lin, S.; Zhu, J.; Dai, F.-H.; Gao, J.; She, J.-L.; Mei, Z.; et al. Gut Microbiome Alterations and Gut Barrier Dysfunction Are Associated with Host Immune Homeostasis in COVID-19 Patients. BMC Med. 2022, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Jiang, H.; Chen, Y.; Gu, S.; Xia, J.; Zhang, H.; Lu, Y.; Yan, R.; Li, L. The Faecal Metabolome in COVID-19 Patients Is Altered and Associated with Clinical Features and Gut Microbes. Anal. Chim. Acta 2021, 1152, 338267. [Google Scholar] [CrossRef] [PubMed]
- Escarcega, R.D.; Honarpisheh, P.; Colpo, G.D.; Ahnstedt, H.W.; Couture, L.; Juneja, S.; Torres, G.; Ortiz, G.J.; Sollome, J.; Tabor, N.; et al. Sex Differences in Global Metabolomic Profiles of COVID-19 Patients. Cell Death Dis. 2022, 13, 461. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Fei, W.; Ye, Y.; Zhao, M.; Zheng, C. The Role of Short-Chain Fatty Acids in Immunity, Inflammation and Metabolism. Crit. Rev. Food Sci. Nutr. 2022, 62, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Xu, C.; Feng, B.; Gao, X.; Liu, Z. Critical Roles of Bile Acids in Regulating Intestinal Mucosal Immune Responses. Ther. Adv. Gastroenterol. 2021, 14, 17562848211018098. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. The Role of Butyrate in Attenuating Pathobiont-Induced Hyperinflammation. Immune Netw. 2020, 20, e15. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-Cell Generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhao, K.-N.; Vitetta, L. Effects of Intestinal Microbial–Elaborated Butyrate on Oncogenic Signaling Pathways. Nutrients 2019, 11, 1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Wang, Y.; Yang, G.; Zhang, Q.; Meng, L.; Xin, Y.; Jiang, X. The Role of Short-Chain Fatty Acids in Intestinal Barrier Function, Inflammation, Oxidative Stress, and Colonic Carcinogenesis. Pharmacol. Res. 2021, 165, 105420. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Takeuchi, T.; Masuoka, H.; Aoki, R.; Ishikane, M.; Iwamoto, N.; Sugiyama, M.; Suda, W.; Nakanishi, Y.; Terada-Hirashima, J.; et al. Human Gut Microbiota and Its Metabolites Impact Immune Responses in COVID-19 and Its Complications. Gastroenterology 2022. [Google Scholar] [CrossRef] [PubMed]
- García-Lafuente, A.; Antolín, M.; Guarner, F.; Crespo, E.; Malagelada, J.-R. Modulation of Colonic Barrier Function by the Composition of the Commensal Flora in the Rat. Gut 2001, 48, 503–507. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Tao, W.; Flavell, R.A.; Zhu, S. Potential Intestinal Infection and Faecal–Oral Transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 269–283. [Google Scholar] [CrossRef]
- Edwinson, A.; Yang, L.; Chen, J.; Grover, M. Colonic Expression of Ace2, the SARS-CoV-2 Entry Receptor, Is Suppressed by Commensal Human Microbiota. Gut Microbes 2021, 13, 1984105. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Sun, Y.; Ren, Z.; Zhu, W.; Li, A.; Cui, G. Potential Associations Between Microbiome and COVID-19. Front. Med. 2021, 8, 785496. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, R.; Zhou, Q. ACE2, B0AT1, and SARS-CoV-2 Spike Protein: Structural and Functional Implications. Curr. Opin. Struct. Biol. 2022, 74, 102388. [Google Scholar] [CrossRef]
- Perlot, T.; Penninger, J.M. ACE2—from the Renin-Angiotensin System to Gut Microbiota and Malnutrition. Microbes Infect. 2013, 15, 866–873. [Google Scholar] [CrossRef]
- He, Y.; Wang, J.; Li, F.; Shi, Y. Main Clinical Features of COVID-19 and Potential Prognostic and Therapeutic Value of the Microbiota in SARS-CoV-2 Infections. Front. Microbiol. 2020, 11, 1302. [Google Scholar] [CrossRef]
- Chen, T.-H.; Hsu, M.-T.; Lee, M.-Y.; Chou, C.-K. Gastrointestinal Involvement in SARS-CoV-2 Infection. Viruses 2022, 14, 1188. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, V.M.; Engel, D.F.; Ricci, M.F.; Cruz, C.S.; Lopes, I.S.; Alves, D.A.; Auriol, M.D.; Magalhães, J.; Zuccoli, G.S.; Smith, B.J.; et al. Gut Microbiota from Patients with Mild COVID-19 Cause Alterations in Mice That Resemble Post-COVID Syndrome. Researchsquare 2022. [Google Scholar] [CrossRef]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The Gut Microbiota Influences Blood-Brain Barrier Permeability in Mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Bi, W.; Xiao, S.; Lan, X.; Cheng, X.; Zhang, J.; Lu, D.; Wei, W.; Wang, Y.; Li, H.; et al. Neuroinflammation Induced by Lipopolysaccharide Causes Cognitive Impairment in Mice. Sci. Rep. 2019, 9, 5790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manosso, L.M.; Arent, C.O.; Borba, L.A.; Ceretta, L.B.; Quevedo, J.; Réus, G.Z. Microbiota-Gut-Brain Communication in the SARS-CoV-2 Infection. Cells 2021, 10, 1993. [Google Scholar] [CrossRef]
- Xu, J.; Wu, Z.; Zhang, M.; Liu, S.; Zhou, L.; Yang, C.; Liu, C. The Role of the Gastrointestinal System in Neuroinvasion by SARS-CoV-2. Front Neurosci. 2021, 15, 694446. [Google Scholar] [CrossRef] [PubMed]
- Pilz, S.; Theiler-Schwetz, V.; Trummer, C.; Krause, R.; Ioannidis, J.P.A. SARS-CoV-2 Reinfections: Overview of Efficacy and Duration of Natural and Hybrid Immunity. Environ. Res. 2022, 209, 112911. [Google Scholar] [CrossRef] [PubMed]
- Cromer, D.; Juno, J.A.; Khoury, D.; Reynaldi, A.; Wheatley, A.K.; Kent, S.J.; Davenport, M.P. Prospects for Durable Immune Control of SARS-CoV-2 and Prevention of Reinfection. Nat. Rev. Immunol. 2021, 21, 395–404. [Google Scholar] [CrossRef]
- Goldblatt, D. SARS-CoV-2: From Herd Immunity to Hybrid Immunity. Nat. Rev. Immunol. 2022, 22, 333–334. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Qi, H.; Liu, B.; Wang, X.; Zhang, L. The Humoral Response and Antibodies against SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 1008–1020. [Google Scholar] [CrossRef]
- Castro Dopico, X.; Ols, S.; Loré, K.; Karlsson Hedestam, G.B. Immunity to SARS-CoV-2 Induced by Infection or Vaccination. J. Intern. Med. 2022, 291, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Leidi, A.; Koegler, F.; Dumont, R.; Dubos, R.; Zaballa, M.-E.; Piumatti, G.; Coen, M.; Berner, A.; Darbellay Farhoumand, P.; Vetter, P.; et al. Risk of Reinfection After Seroconversion to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): A Population-Based Propensity-Score Matched Cohort Study. Clin. Infect. Dis. 2022, 74, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Finch, E.; Lowe, R.; Fischinger, S.; Aubin, M.d.S.; Siddiqui, S.M.; Dayal, D.; Loesche, M.A.; Rhee, J.; Beger, S.; Hu, Y.; et al. SARS-CoV-2 Antibodies Protect against Reinfection for at Least 6 Months in a Multicentre Seroepidemiological Workplace Cohort. PLOS Biol. 2022, 20, e3001531. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Muecksch, F.; Schaefer-Babajew, D.; Finkin, S.; Viant, C.; Gaebler, C.; Hoffmann, H.-H.; Barnes, C.O.; Cipolla, M.; Ramos, V.; et al. Naturally Enhanced Neutralizing Breadth against SARS-CoV-2 One Year after Infection. Nature 2021, 595, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection from Symptomatic SARS-CoV-2 Infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and Cellular Immune Memory to Four COVID-19 Vaccines. Cell 2022, 185, 2434–2451. [Google Scholar] [CrossRef]
- Bertoletti, A.; Le Bert, N.; Qui, M.; Tan, A.T. SARS-CoV-2-Specific T Cells in Infection and Vaccination. Cell. Mol. Immunol. 2021, 18, 2307–2312. [Google Scholar] [CrossRef] [PubMed]
- Epsi, N.J.; Richard, S.A.; Lindholm, D.A.; Mende, K.; Ganesan, A.; Huprikar, N.; Lalani, T.; Fries, A.C.; Maves, R.C.; Colombo, R.E.; et al. Understanding ‘Hybrid Immunity’: Comparison and Predictors of Humoral Immune Responses to SARS-CoV-2 Infection and COVID-19 Vaccines. Clin. Infect. Dis. 2022, ciac392. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Z.; Azman, A.S.; Sun, R.; Lu, W.; Zheng, N.; Zhou, J.; Wu, Q.; Deng, X.; Zhao, Z.; et al. Neutralizing Antibodies Against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants Induced by Natural Infection or Vaccination: A Systematic Review and Pooled Analysis. Clin. Infect. Dis. 2022, 74, 734–742. [Google Scholar] [CrossRef]
- Goldberg, Y.; Mandel, M.; Woodbridge, Y.; Fluss, R.; Novikov, I.; Yaari, R.; Ziv, A.; Freedman, L.; Huppert, A. Similarity of Protection Conferred by Previous SARS-CoV-2 Infection and by BNT162b2 Vaccine: A 3-Month Nationwide Experience from Israel. Am. J. Epidemiol. 2022, 191, kwac060. [Google Scholar] [CrossRef] [PubMed]
- Chemaitelly, H.; Ayoub, H.H.; AlMukdad, S.; Coyle, P.; Tang, P.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Hasan, M.R.; Al-Kanaani, Z.; et al. Protection from Previous Natural Infection Compared with MRNA Vaccination against SARS-CoV-2 Infection and Severe COVID-19 in Qatar: A Retrospective Cohort Study. Lancet Microbe 2022, 3, e944–e955. [Google Scholar] [CrossRef] [PubMed]
- Bates, T.A.; McBride, S.K.; Leier, H.C.; Guzman, G.; Lyski, Z.L.; Schoen, D.; Winders, B.; Lee, J.-Y.; Lee, D.X.; Messer, W.B.; et al. Vaccination before or after SARS-CoV-2 Infection Leads to Robust Humoral Response and Antibodies That Effectively Neutralize Variants. Sci. Immunol. 2022, 7, eabn8014. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.I.; Baskett, W.I.; Huang, W.; Lobanova, I.; Hasan Naqvi, S.; Shyu, C.-R. Reinfection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Patients Undergoing Serial Laboratory Testing. Clin. Infect. Dis. 2022, 74, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Abu-Raddad, L.J.; Chemaitelly, H.; Malek, J.A.; Ahmed, A.A.; Mohamoud, Y.A.; Younuskunju, S.; Ayoub, H.H.; Al Kanaani, Z.; Al Khal, A.; Al Kuwari, E.; et al. Assessment of the Risk of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Reinfection in an Intense Reexposure Setting. Clin. Infect. Dis. 2021, 73, e1830–e1840. [Google Scholar] [CrossRef] [PubMed]
- Abu-Raddad, L.J.; Chemaitelly, H.; Coyle, P.; Malek, J.A.; Ahmed, A.A.; Mohamoud, Y.A.; Younuskunju, S.; Ayoub, H.H.; Al Kanaani, Z.; Al Kuwari, E.; et al. SARS-CoV-2 Antibody-Positivity Protects against Reinfection for at Least Seven Months with 95% Efficacy. EClinicalMedicine 2021, 35, 100861. [Google Scholar] [CrossRef] [PubMed]
- Abu-Raddad, L.J.; Chemaitelly, H.; Bertollini, R. Severity of SARS-CoV-2 Reinfections as Compared with Primary Infections. N. Engl. J. Med. 2021, 385, 2487–2489. [Google Scholar] [CrossRef]
- Mensah, A.A.; Lacy, J.; Stowe, J.; Seghezzo, G.; Sachdeva, R.; Simmons, R.; Bukasa, A.; O’Boyle, S.; Andrews, N.; Ramsay, M.; et al. Disease Severity during SARS-COV-2 Reinfection: A Nationwide Study. J. Infect. 2022, 84, 542–550. [Google Scholar] [CrossRef]
- Mohsin, M.; Mahmud, S. Omicron SARS-CoV-2 Variant of Concern: A Review on Its Transmissibility, Immune Evasion, Reinfection, and Severity. Medicine (Baltimore) 2022, 101, e29165. [Google Scholar] [CrossRef]
- Nguyen, N.N.; Houhamdi, L.; Hoang, V.T.; Delerce, J.; Delorme, L.; Colson, P.; Brouqui, P.; Fournier, P.-E.; Raoult, D.; Gautret, P. SARS-CoV-2 Reinfection and COVID-19 Severity. Emerg. Microbes Infect. 2022, 11, 894–901. [Google Scholar] [CrossRef]
- Pulliam, J.R.C.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased Risk of SARS-CoV-2 Reinfection Associated with Emergence of Omicron in South Africa. Science 2022, 376, eabn4947. [Google Scholar] [CrossRef]
- Altarawneh, H.N.; Chemaitelly, H.; Hasan, M.R.; Ayoub, H.H.; Qassim, S.; AlMukdad, S.; Coyle, P.; Yassine, H.M.; Al-Khatib, H.A.; Benslimane, F.M.; et al. Protection against the Omicron Variant from Previous SARS-CoV-2 Infection. N. Engl. J. Med. 2022, 386, 1288–1290. [Google Scholar] [CrossRef] [PubMed]
- Qassim, S.H.; Chemaitelly, H.; Ayoub, H.H.; AlMukdad, S.; Tang, P.; Hasan, M.R.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Abdul-Rahim, H.F.; et al. Effects of BA.1/BA.2 Subvariant, Vaccination and Prior Infection on Infectiousness of SARS-CoV-2 Omicron Infections. J. Travel Med. 2022, 29, taac068. [Google Scholar] [CrossRef] [PubMed]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.J.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C.; et al. Clinical Severity of SARS-CoV-2 Omicron BA.4 and BA.5 Lineages Compared to BA.1 and Delta in South Africa. Nat. Commun. 2022, 13, 5860. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, N. Report 50: Hospitalisation Risk for Omicron Cases in England; Imperial College London: London, UK, 2021. [Google Scholar]
- Medić, S.; Anastassopoulou, C.; Lozanov-Crvenković, Z.; Vuković, V.; Dragnić, N.; Petrović, V.; Ristić, M.; Pustahija, T.; Gojković, Z.; Tsakris, A.; et al. Risk and Severity of SARS-CoV-2 Reinfections during 2020–2022 in Vojvodina, Serbia: A Population-Level Observational Study. Lancet Reg. Health—Eur. 2022, 20, 100453. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Zamora, E.; Mendoza-Cano, O.; Delgado-Enciso, I.; Hernandez-Suarez, C.M. Predictors of Severe Symptomatic Laboratory-Confirmed SARS-CoV-2 Reinfection. Public Health 2021, 193, 113–115. [Google Scholar] [CrossRef]
- Islam, M.Z.; Riaz, B.K.; Akbar Ashrafi, S.A.; Farjana, S.; Efa, S.S.; Khan, M.A. Severity of COVID-19 Reinfection and Associated Risk Factors: Findings of a Cross-Sectional Study in Bangladesh; Infectious Diseases (except HIV/AIDS). medRxiv 2022. [Google Scholar] [CrossRef]
- Comba, I.Y.; Riestra Guiance, I.; Corsini Campioli, C.; Challener, D.; Sampathkumar, P.; Orenstein, R.; Gordon, J.; Bosch, W.; O’Horo, J.C. Clinical Characteristics and Outcomes of Patients with SARS-CoV-2 Reinfection. Mayo Clin. Proc. Innov. Qual. Outcomes 2022, 6, 361–372. [Google Scholar] [CrossRef]
- Guthmiller, J.J.; Stovicek, O.; Wang, J.; Changrob, S.; Li, L.; Halfmann, P.; Zheng, N.-Y.; Utset, H.; Stamper, C.T.; Dugan, H.L.; et al. SARS-CoV-2 Infection Severity Is Linked to Superior Humoral Immunity against the Spike. mBio 2021, 12, e02940-20. [Google Scholar] [CrossRef]
- Trinité, B.; Tarrés-Freixas, F.; Rodon, J.; Pradenas, E.; Urrea, V.; Marfil, S.; Rodríguez de la Concepción, M.L.; Ávila-Nieto, C.; Aguilar-Gurrieri, C.; Barajas, A.; et al. SARS-CoV-2 Infection Elicits a Rapid Neutralizing Antibody Response That Correlates with Disease Severity. Sci. Rep. 2021, 11, 2608. [Google Scholar] [CrossRef]
- Laing, E.D.; Epsi, N.J.; Richard, S.A.; Samuels, E.C.; Wang, W.; Vassell, R.; Ewing, D.F.; Herrup, R.; Sterling, S.L.; Lindholm, D.A.; et al. SARS-CoV-2 Antibodies Remain Detectable 12 Months after Infection and Antibody Magnitude Is Associated with Age and COVID-19 Severity. medRxiv 2021. [Google Scholar] [CrossRef]
- Legros, V.; Denolly, S.; Vogrig, M.; Boson, B.; Siret, E.; Rigaill, J.; Pillet, S.; Grattard, F.; Gonzalo, S.; Verhoeven, P.; et al. A Longitudinal Study of SARS-CoV-2-Infected Patients Reveals a High Correlation between Neutralizing Antibodies and COVID-19 Severity. Cell. Mol. Immunol. 2021, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Manuylov, V.; Burgasova, O.; Borisova, O.; Smetanina, S.; Vasina, D.; Grigoriev, I.; Kudryashova, A.; Semashko, M.; Cherepovich, B.; Kharchenko, O.; et al. Avidity of IgG to SARS-CoV-2 RBD as a Prognostic Factor for the Severity of COVID-19 Reinfection. Viruses 2022, 14, 617. [Google Scholar] [CrossRef] [PubMed]
- Wat, D. The Common Cold: A Review of the Literature. Eur. J. Intern. Med. 2004, 15, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Seiler, P.; Jones, J.C.; Ridout, G.; Camp, K.P.; Fabrizio, T.P.; Jeevan, T.; Miller, L.A.; Throm, R.E.; Ferrara, F.; et al. Antibody Responses to SARS-CoV-2 Antigens in Humans and Animals. Vaccines 2020, 8, 684. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.C.S.; Owen, C.J.; Tham, C.Y.L.; Bertoletti, A.; van Dorp, L.; Balloux, F. Pre-Existing T Cell-Mediated Cross-Reactivity to SARS-CoV-2 Cannot Solely Be Explained by Prior Exposure to Endemic Human Coronaviruses. Infect. Genet. Evol. 2021, 95, 105075. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Kode, V.; Bhojak, K.; Karunakaran, C.; Lee, K.; Manoharan, M.; Ramesh, A.; Hv, S.; Srivastava, A.; Sathian, R.; et al. Immunodominant T-Cell Epitopes from the SARS-CoV-2 Spike Antigen Reveal Robust Pre-Existing T-Cell Immunity in Unexposed Individuals. Sci. Rep. 2021, 11, 13164. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-Reactive T Cells in Healthy Donors and Patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-Specific T Cell Immunity in Cases of COVID-19 and SARS, and Uninfected Controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Becerra-Artiles, A.; Calvo-Calle, J.M.; Co, M.D.; Nanaware, P.P.; Cruz, J.; Weaver, G.C.; Lu, L.; Forconi, C.; Finberg, R.W.; Moormann, A.M.; et al. Broadly Recognized, Cross-Reactive SARS-CoV-2 CD4 T Cell Epitopes Are Highly Conserved across Human Coronaviruses and Presented by Common HLA Alleles. Cell Rep. 2022, 39, 110952. [Google Scholar] [CrossRef]
- Weiskopf, D. SARS-CoV-2 Specific and Cross-Reactive t Cell Responses. Top. Antivir. Med. 2021, 29, 4. [Google Scholar]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; Sette, A.; et al. Single-Cell Transcriptomic Analysis of SARS-CoV-2 Reactive CD4+ T Cells. Cell 2020, 183, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.; Arya, R.; Sachan, S.; Jha, S.N.; Kalia, A.; Lall, A.; Sette, A.; Grifoni, A.; Weiskopf, D.; Coshic, P.; et al. Immune Memory in Mild COVID-19 Patients and Unexposed Donors Reveals Persistent T Cell Responses After SARS-CoV-2 Infection. Front. Immunol. 2021, 12, 749. [Google Scholar] [CrossRef] [PubMed]
- Dykema, A.G.; Zhang, B.; Woldemeskel, B.A.; Garliss, C.C.; Cheung, L.S.; Choudhury, D.; Zhang, J.; Aparicio, L.; Bom, S.; Rashid, R.; et al. Functional Characterization of CD4+ T Cell Receptors Crossreactive for SARS-CoV-2 and Endemic Coronaviruses. J. Clin. Investig. 2021, 131, e146922. [Google Scholar] [CrossRef] [PubMed]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-Derived Peptides Define Heterologous and COVID-19-Induced T Cell Recognition. Nat. Immunol. 2021, 22, 74–85. [Google Scholar] [CrossRef]
- Schulien, I.; Kemming, J.; Oberhardt, V.; Wild, K.; Seidel, L.M.; Killmer, S.; Sagar; Daul, F.; Salvat Lago, M.; Decker, A.; et al. Characterization of Pre-Existing and Induced SARS-CoV-2-Specific CD8+ T Cells. Nat. Med. 2021, 27, 78–85. [Google Scholar] [CrossRef]
- Schmidt, K.G.; Nganou-Makamdop, K.; Tenbusch, M.; El Kenz, B.; Maier, C.; Lapuente, D.; Überla, K.; Spriewald, B.; Bergmann, S.; Harrer, E.G.; et al. SARS-CoV-2-Seronegative Subjects Target CTL Epitopes in the SARS-CoV-2 Nucleoprotein Cross-Reactive to Common Cold Coronaviruses. Front. Immunol. 2021, 12, 627568. [Google Scholar] [CrossRef]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and Strong Memory CD4+ and CD8+ T Cells Induced by SARS-CoV-2 in UK Convalescent Individuals Following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Lineburg, K.E.; Grant, E.J.; Swaminathan, S.; Chatzileontiadou, D.S.M.; Szeto, C.; Sloane, H.; Panikkar, A.; Raju, J.; Crooks, P.; Rehan, S.; et al. CD8+ T Cells Specific for an Immunodominant SARS-CoV-2 Nucleocapsid Epitope Cross-React with Selective Seasonal Coronaviruses. Immunity 2021, 54, 1055–1065.e5. [Google Scholar] [CrossRef]
- Loyal, L.; Braun, J.; Henze, L.; Kruse, B.; Dingeldey, M.; Reimer, U.; Kern, F.; Schwarz, T.; Mangold, M.; Unger, C.; et al. Cross-Reactive CD4+ T Cells Enhance SARS-CoV-2 Immune Responses upon Infection and Vaccination. Science 2021, 374, eabh1823. [Google Scholar] [CrossRef]
- Lipsitch, M.; Grad, Y.H.; Sette, A.; Crotty, S. Cross-Reactive Memory T Cells and Herd Immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020, 20, 709–713. [Google Scholar] [CrossRef]
- Moss, P. The T Cell Immune Response against SARS-CoV-2. Nat. Immunol. 2022, 23, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Majdoubi, A.; Michalski, C.; O’Connell, S.E.; Dada, S.; Narpala, S.; Gelinas, J.; Mehta, D.; Cheung, C.; Winkler, D.F.H.; Basappa, M.; et al. A Majority of Uninfected Adults Show Preexisting Antibody Reactivity against SARS-CoV-2. JCI Insight 2021, 6, e146316. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.R.; Steiner, D.; Pihl, R.M.F.; Chavez, E.; Olson, A.; Smith, E.L.; Baird, L.A.; Korkmaz, F.; Urick, P.; Sagar, M.; et al. Novel ELISA Protocol Links Pre-Existing SARS-CoV-2 Reactive Antibodies with Endemic Coronavirus Immunity and Age and Reveals Improved Serologic Identification of Acute COVID-19 via Multi-Parameter Detection. Front. Immunol. 2021, 12, 614676. [Google Scholar] [CrossRef] [PubMed]
- Dalakas, M.C.; Bitzogli, K.; Alexopoulos, H. Anti-SARS-CoV-2 Antibodies within IVIg Preparations: Cross-Reactivities with Seasonal Coronaviruses, Natural Autoimmunity, and Therapeutic Implications. Front. Immunol. 2021, 12, 627285. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Y.; Kang, L.; Hu, Y.; Wang, L.; Zhong, J.; Chen, H.; Ren, L.; Gu, X.; Wang, G.; et al. Cross-Reactive Antibody against Human Coronavirus OC43 Spike Protein Correlates with Disease Severity in COVID-19 Patients: A Retrospective Study. Emerg. Microbes Infect. 2021, 10, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Kubota-Koketsu, R.; Terada, Y.; Yunoki, M.; Sasaki, T.; Nakayama, E.E.; Kamitani, W.; Shioda, T. Neutralizing and Binding Activities against SARS-CoV-1/2, MERS-CoV, and Human Coronaviruses 229E and OC43 by Normal Human Intravenous Immunoglobulin Derived from Healthy Donors in Japan. Transfusion 2021, 61, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.E.; Tan, C.W.; Chia, W.N.; Young, B.E.; Linster, M.; Low, J.H.; Tan, Y.-J.; Chen, M.I.-C.; Smith, G.J.D.; Leo, Y.S.; et al. Lack of Cross-Neutralization by SARS Patient Sera towards SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 900–902. [Google Scholar] [CrossRef]
- García-Jiménez, Á.F.; Cáceres-Martell, Y.; Fernández-Soto, D.; Martínez Fleta, P.; Casasnovas, J.M.; Sánchez-Madrid, F.; Frade, J.M.R.; Valés-Gómez, M.; Reyburn, H.T. Cross-Reactive Cellular, but Not Humoral, Immunity Is Detected between OC43 and SARS-CoV-2 NPs in People Not Infected with SARS-CoV-2: Possible Role of CTFH Cells. J. Leukoc. Biol. 2022, 112, 339–346. [Google Scholar] [CrossRef]
- Song, G.; He, W.; Callaghan, S.; Anzanello, F.; Huang, D.; Ricketts, J.; Torres, J.L.; Beutler, N.; Peng, L.; Vargas, S.; et al. Cross-Reactive Serum and Memory B-Cell Responses to Spike Protein in SARS-CoV-2 and Endemic Coronavirus Infection. Nat. Commun. 2021, 12, 2938. [Google Scholar] [CrossRef]
- Hicks, J.; Klumpp-Thomas, C.; Kalish, H.; Shunmugavel, A.; Mehalko, J.; Denson, J.-P.; Snead, K.R.; Drew, M.; Corbett, K.S.; Graham, B.S.; et al. Serologic Cross-Reactivity of SARS-CoV-2 with Endemic and Seasonal Betacoronaviruses. J. Clin. Immunol. 2021, 41, 906–913. [Google Scholar] [CrossRef]
- Tso, F.Y.; Lidenge, S.J.; Peña, P.B.; Clegg, A.A.; Ngowi, J.R.; Mwaiselage, J.; Ngalamika, O.; Julius, P.; West, J.T.; Wood, C. High Prevalence of Pre-Existing Serological Cross-Reactivity against Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in Sub-Saharan Africa. Int. J. Infect. Dis. 2021, 102, 577–583. [Google Scholar] [CrossRef]
- Tajuelo, A.; López-Siles, M.; Más, V.; Pérez-Romero, P.; Aguado, J.M.; Briz, V.; McConnell, M.J.; Martín-Galiano, A.J.; López, D. Cross-Recognition of SARS-CoV-2 B-Cell Epitopes with Other Betacoronavirus Nucleoproteins. Int. J. Mol. Sci. 2022, 23, 2977. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, C.I.; Sung, K.; Ojee, E.; Adhiambo, J.; Begnel, E.R.; Slyker, J.; Gantt, S.; Matsen, F.A.; Kinuthia, J.; Wamalwa, D.; et al. Distinct Antibody Responses to Endemic Coronaviruses Pre- and Post-SARS-CoV-2 Infection in Kenyan Infants and Mothers. Viruses 2022, 14, 1517. [Google Scholar] [CrossRef] [PubMed]
- Denninger, V.; Xu, C.K.; Meisl, G.; Morgunov, A.S.; Fiedler, S.; Ilsley, A.; Emmenegger, M.; Malik, A.Y.; Piziorska, M.A.; Schneider, M.M.; et al. Understanding the Role of Memory Re-Activation and Cross-Reactivity in the Defense against SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J.; et al. Preexisting and de Novo Humoral Immunity to SARS-CoV-2 in Humans. Science 2020, 370, 1339–1343. [Google Scholar] [CrossRef] [PubMed]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.-H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral Epitope Profiling of COVID-19 Patients Reveals Cross-Reactivity and Correlates of Severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Young, B.E.; Li, D.; Seppo, A.; Zhou, Q.; Wiltse, A.; Nowak-Wegrzyn, A.; Murphy, K.; Widrick, K.; Diaz, N.; et al. Broad Cross-Reactive IgA and IgG against Human Coronaviruses in Milk Induced by COVID-19 Vaccination and Infection. Vaccines 2022, 10, 980. [Google Scholar] [CrossRef]
- Aydillo, T.; Rombauts, A.; Stadlbauer, D.; Aslam, S.; Abelenda-Alonso, G.; Escalera, A.; Amanat, F.; Jiang, K.; Krammer, F.; Carratala, J.; et al. Immunological Imprinting of the Antibody Response in COVID-19 Patients. Nat. Commun. 2021, 12, 3781. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.A.; Kellogg, C.; Equils, O. Neutralizing and Cross-Reacting Antibodies: Implications for Immunotherapy and SARS-CoV-2 Vaccine Development. Hum. Vaccines Immunother. 2021, 17, 84–87. [Google Scholar] [CrossRef]
- Aguilar-Bretones, M.; Westerhuis, B.M.; Raadsen, M.P.; Bruin, E.D.; Chandler, F.D.; Okba, N.M.A.; Haagmans, B.L.; Langerak, T.; Endeman, H.; van den Akker, J.P.C.; et al. Seasonal Coronavirus–Specific B Cells with Limited SARS-CoV-2 Cross-Reactivity Dominate the IgG Response in Severe COVID-19. J. Clin. Investig. 2021, 131, e150613. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Wolf, J.; Brice, D.C.; Sun, Y.; Locke, M.; Cherry, S.; Castellaw, A.H.; Wehenkel, M.; Crawford, J.C.; Zarnitsyna, V.I.; et al. Pre-Existing Humoral Immunity to Human Common Cold Coronaviruses Negatively Impacts the Protective SARS-CoV-2 Antibody Response. Cell Host Microbe 2022, 30, 83–96.e4. [Google Scholar] [CrossRef]
- Anderson, E.M.; Goodwin, E.C.; Verma, A.; Arevalo, C.P.; Bolton, M.J.; Weirick, M.E.; Gouma, S.; McAllister, C.M.; Christensen, S.R.; Weaver, J.; et al. Seasonal Human Coronavirus Antibodies Are Boosted upon SARS-CoV-2 Infection but Not Associated with Protection. Cell 2021, 184, 1858–1864.e10. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Contant, P.; Embong, A.K.; Kanagaiah, P.; Chaves, F.A.; Yang, H.; Branche, A.R.; Topham, D.J.; Sangster, M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. mBio 2020, 11, e01991-20. [Google Scholar] [CrossRef] [PubMed]
- Prévost, J.; Gasser, R.; Beaudoin-Bussières, G.; Richard, J.; Duerr, R.; Laumaea, A.; Anand, S.P.; Goyette, G.; Benlarbi, M.; Ding, S.; et al. Cross-Sectional Evaluation of Humoral Responses against SARS-CoV-2 Spike. Cell Rep. Med. 2020, 1, 100126. [Google Scholar] [CrossRef] [PubMed]
- Crowley, A.R.; Natarajan, H.; Hederman, A.P.; Bobak, C.A.; Weiner, J.A.; Wieland-Alter, W.; Lee, J.; Bloch, E.M.; Tobian, A.A.; Redd, A.D.; et al. Boosting of Cross-Reactive Antibodies to Endemic Coronaviruses by SARS-CoV-2 Infection but Not Vaccination with Stabilized Spike. eLife 2022, 11, e75228. [Google Scholar] [CrossRef]
- Becker, M.; Strengert, M.; Junker, D.; Kaiser, P.D.; Kerrinnes, T.; Traenkle, B.; Dinter, H.; Häring, J.; Ghozzi, S.; Zeck, A.; et al. Exploring beyond Clinical Routine SARS-CoV-2 Serology Using MultiCoV-Ab to Evaluate Endemic Coronavirus Cross-Reactivity. Nat. Commun. 2021, 12, 1152. [Google Scholar] [CrossRef] [PubMed]
- Ortega, N.; Ribes, M.; Vidal, M.; Rubio, R.; Aguilar, R.; Williams, S.; Barrios, D.; Alonso, S.; Hernández-Luis, P.; Mitchell, R.A.; et al. Seven-Month Kinetics of SARS-CoV-2 Antibodies and Role of Pre-Existing Antibodies to Human Coronaviruses. Nat. Commun. 2021, 12, 4740. [Google Scholar] [CrossRef]
- Geanes, E.S.; LeMaster, C.; Fraley, E.R.; Khanal, S.; McLennan, R.; Grundberg, E.; Selvarangan, R.; Bradley, T. Cross-Reactive Antibodies Elicited to Conserved Epitopes on SARS-CoV-2 Spike Protein after Infection and Vaccination. Sci. Rep. 2022, 12, 6496. [Google Scholar] [CrossRef]
- Dugas, M.; Grote-Westrick, T.; Vollenberg, R.; Lorentzen, E.; Brix, T.; Schmidt, H.; Tepasse, P.-R.; Kühn, J. Less Severe Course of COVID-19 Is Associated with Elevated Levels of Antibodies against Seasonal Human Coronaviruses OC43 and HKU1 (HCoV OC43, HCoV HKU1). Int. J. Infect. Dis. 2021, 105, 304–306. [Google Scholar] [CrossRef]
- Dugas, M.; Grote-Westrick, T.; Merle, U.; Fontenay, M.; Kremer, A.E.; Hanses, F.; Vollenberg, R.; Lorentzen, E.; Tiwari-Heckler, S.; Duchemin, J.; et al. Lack of Antibodies against Seasonal Coronavirus OC43 Nucleocapsid Protein Identifies Patients at Risk of Critical COVID-19. J. Clin. Virol. 2021, 139, 104847. [Google Scholar] [CrossRef]
- Henss, L.; Scholz, T.; von Rhein, C.; Wieters, I.; Borgans, F.; Eberhardt, F.J.; Zacharowski, K.; Ciesek, S.; Rohde, G.; Vehreschild, M.; et al. Analysis of Humoral Immune Responses in Patients with Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Infect. Dis. 2021, 223, 56–61. [Google Scholar] [CrossRef]
- Sagar, M.; Reifler, K.; Rossi, M.; Miller, N.S.; Sinha, P.; White, L.F.; Mizgerd, J.P. Recent Endemic Coronavirus Infection Is Associated with Less-Severe COVID-19. J. Clin. Investig. 2021, 131, e143380. [Google Scholar] [CrossRef]
- Kaplonek, P.; Wang, C.; Bartsch, Y.; Fischinger, S.; Gorman, M.J.; Bowman, K.; Kang, J.; Dayal, D.; Martin, P.; Nowak, R.P.; et al. Early Cross-Coronavirus Reactive Signatures of Humoral Immunity against COVID-19. Sci. Immunol. 2021, 6, eabj2901. [Google Scholar] [CrossRef] [PubMed]
- Gombar, S.; Bergquist, T.; Pejaver, V.; Hammarlund, N.E.; Murugesan, K.; Mooney, S.; Shah, N.; Pinsky, B.A.; Banaei, N. SARS-CoV-2 Infection and COVID-19 Severity in Individuals with Prior Seasonal Coronavirus Infection. Diagn. Microbiol. Infect. Dis. 2021, 100, 115338. [Google Scholar] [CrossRef] [PubMed]
- Loos, C.; Atyeo, C.; Fischinger, S.; Burke, J.; Slein, M.D.; Streeck, H.; Lauffenburger, D.; Ryan, E.T.; Charles, R.C.; Alter, G. Evolution of Early SARS-CoV-2 and Cross-Coronavirus Immunity. mSphere 2020, 5, e00622-20. [Google Scholar] [CrossRef] [PubMed]
- Dispinseri, S.; Marzinotto, I.; Brigatti, C.; Pirillo, M.F.; Tolazzi, M.; Bazzigaluppi, E.; Canitano, A.; Borghi, M.; Gallinaro, A.; Caccia, R.; et al. Seasonal Betacoronavirus Antibodies’ Expansion Post-BNT161b2 Vaccination Associates with Reduced SARS-CoV-2 VoC Neutralization. J. Clin. Immunol. 2022, 42, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Tetro, J.A. Is COVID-19 Receiving ADE from Other Coronaviruses? Microbes Infect. 2020, 22, 72–73. [Google Scholar] [CrossRef]
- Borrega, R.; Nelson, D.K.S.; Koval, A.P.; Bond, N.G.; Heinrich, M.L.; Rowland, M.M.; Lathigra, R.; Bush, D.J.; Aimukanova, I.; Phinney, W.N.; et al. Cross-Reactive Antibodies to SARS-CoV-2 and MERS-CoV in Pre-COVID-19 Blood Samples from Sierra Leoneans. Viruses 2021, 13, 2325. [Google Scholar] [CrossRef]
- Galipeau, Y.; Siragam, V.; Laroche, G.; Marion, E.; Greig, M.; McGuinty, M.; Booth, R.A.; Durocher, Y.; Cuperlovic-Culf, M.; Bennett, S.A.L.; et al. Relative Ratios of Human Seasonal Coronavirus Antibodies Predict the Efficiency of Cross-Neutralization of SARS-CoV-2 Spike Binding to ACE2. eBioMedicine 2021, 74, 103700. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Wells, D.A.; Cantoni, D.; Mayora-Neto, M.; Di Genova, C.; Sampson, A.; Ferrari, M.; Carnell, G.; Nadesalingam, A.; Smith, P.; Chan, A.; et al. Human Seasonal Coronavirus Neutralisation and COVID-19 Severity. J. Med. Virol. 2022, 94, 4820–4829. [Google Scholar] [CrossRef]
- Poston, D.; Weisblum, Y.; Wise, H.; Templeton, K.; Jenks, S.; Hatziioannou, T.; Bieniasz, P. Absence of Severe Acute Respiratory Syndrome Coronavirus 2 Neutralizing Activity in Prepandemic Sera from Individuals with Recent Seasonal Coronavirus Infection. Clin. Infect. Dis. 2021, 73, e1208–e1211. [Google Scholar] [CrossRef]
- Schwaiger, J.; Karbiener, M.; Aberham, C.; Farcet, M.R.; Kreil, T.R. No SARS-CoV-2 Neutralization by Intravenous Immunoglobulins Produced from Plasma Collected Before the 2020 Pandemic. J. Infect. Dis. 2020, 222, 1960–1964. [Google Scholar] [CrossRef]
- Sallis, R.; Young, D.R.; Tartof, S.Y.; Sallis, J.F.; Sall, J.; Li, Q.; Smith, G.N.; Cohen, D.A. Physical Inactivity Is Associated with a Higher Risk for Severe COVID-19 Outcomes: A Study in 48 440 Adult Patients. Br. J. Sports Med. 2021, 55, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Lee, J.; Moon, S.Y.; Jin, H.Y.; Yang, J.M.; Ogino, S.; Song, M.; Hong, S.H.; Ghayda, R.A.; Kronbichler, A.; et al. Physical Activity and the Risk of SARS-CoV-2 Infection, Severe COVID-19 Illness and COVID-19 Related Mortality in South Korea: A Nationwide Cohort Study. Br. J. Sports Med. 2022, 56, 901–912. [Google Scholar] [CrossRef]
- Salgado-Aranda, R.; Pérez-Castellano, N.; Núñez-Gil, I.; Orozco, A.J.; Torres-Esquivel, N.; Flores-Soler, J.; Chamaisse-Akari, A.; Mclnerney, A.; Vergara-Uzcategui, C.; Wang, L.; et al. Influence of Baseline Physical Activity as a Modifying Factor on COVID-19 Mortality: A Single-Center, Retrospective Study. Infect. Dis. Ther. 2021, 10, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.-H.; Lee, S.J.; Jae, S.Y.; Kim, W.J.; Ha, S.J.; Gwon, J.G.; Choi, J.; Kim, D.W.; Kim, J.Y. Physical Activity and the Risk of COVID-19 Infection and Mortality: A Nationwide Population-Based Case-Control Study. J. Clin. Med. 2021, 10, 1539. [Google Scholar] [CrossRef] [PubMed]
- Brawner, C.A.; Ehrman, J.K.; Bole, S.; Kerrigan, D.J.; Parikh, S.S.; Lewis, B.K.; Gindi, R.M.; Keteyian, C.; Abdul-Nour, K.; Keteyian, S.J. Inverse Relationship of Maximal Exercise Capacity to Hospitalization Secondary to Coronavirus Disease 2019. Mayo Clin. Proc. 2021, 96, 32–39. [Google Scholar] [CrossRef]
- Ekblom-Bak, E.; Väisänen, D.; Ekblom, B.; Blom, V.; Kallings, L.V.; Hemmingsson, E.; Andersson, G.; Wallin, P.; Salier Eriksson, J.; Holmlund, T.; et al. Cardiorespiratory Fitness and Lifestyle on Severe COVID-19 Risk in 279,455 Adults: A Case Control Study. Int. J. Behav. Nutr. Phys. Act. 2021, 18, 135. [Google Scholar] [CrossRef]
- Christensen, R.A.G.; Arneja, J.; Cyr, K.S.; Sturrock, S.L.; Brooks, J.D. The Association of Estimated Cardiorespiratory Fitness with COVID-19 Incidence and Mortality: A Cohort Study. PLoS ONE 2021, 16, e0250508. [Google Scholar] [CrossRef]
- af Geijerstam, A.; Mehlig, K.; Börjesson, M.; Robertson, J.; Nyberg, J.; Adiels, M.; Rosengren, A.; Åberg, M.; Lissner, L. Fitness, Strength and Severity of COVID-19: A Prospective Register Study of 1 559 187 Swedish Conscripts. BMJ Open 2021, 11, e051316. [Google Scholar] [CrossRef]
- Wong, C.-M.; Lai, H.-K.; Ou, C.-Q.; Ho, S.-Y.; Chan, K.-P.; Thach, T.-Q.; Yang, L.; Chau, Y.-K.; Lam, T.-H.; Hedley, A.J.; et al. Is Exercise Protective Against Influenza-Associated Mortality? PLoS ONE 2008, 3, e2108. [Google Scholar] [CrossRef] [Green Version]
- Matthews, C.E.; Ockene, I.S.; Freedson, P.S.; Rosal, M.C.; Merriam, P.A.; Hebert, J.R. Moderate to Vigorous Physical Activity and Risk of Upper-Respiratory Tract Infection. Med. Sci. Sports Exerc. 2002, 34, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Olivo, C.R.; Miyaji, E.N.; Oliveira, M.L.S.; Almeida, F.M.; Lourenço, J.D.; Abreu, R.M.; Arantes, P.M.M.; Lopes, F.D.; Martins, M.A. Aerobic Exercise Attenuates Pulmonary Inflammation Induced by Streptococcus Pneumoniae. J. Appl. Physiol. 2014, 117, 998–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, J.Y. The Role of Exercise-Induced Myokines in Regulating Metabolism. Arch. Pharm. Res. 2018, 41, 14–29. [Google Scholar] [CrossRef]
- Bruunsgaard, H. Physical Activity and Modulation of Systemic Low-Level Inflammation. J. Leukoc. Biol. 2005, 78, 819–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woods, J.A.; Vieira, V.J.; Keylock, K.T. Exercise, Inflammation, and Innate Immunity. Immunol. Allergy Clin. N. Am. 2009, 29, 381–393. [Google Scholar] [CrossRef]
- Spielmann, G.; McFarlin, B.K.; O’Connor, D.P.; Smith, P.J.W.; Pircher, H.; Simpson, R.J. Aerobic Fitness Is Associated with Lower Proportions of Senescent Blood T-Cells in Man. Brain. Behav. Immun. 2011, 25, 1521–1529. [Google Scholar] [CrossRef]
- Simpson, R.J.; Kunz, H.; Agha, N.; Graff, R. Chapter Fifteen—Exercise and the Regulation of Immune Functions. In Progress in Molecular Biology and Translational Science; Bouchard, C., Ed.; Molecular and Cellular Regulation of Adaptation to Exercise; Academic Press: Cambridge, MA, USA, 2015; Volume 135, pp. 355–380. [Google Scholar]
- Collie, S.; Saggers, R.T.; Bandini, R.; Steenkamp, L.; Champion, J.; Gray, G.; Bekker, L.-G.; Goga, A.; Garrett, N.; Patricios, J. Association between Regular Physical Activity and the Protective Effect of Vaccination against SARS-CoV-2 in a South African Case–Control Study. Br. J. Sports Med. 2022. [Google Scholar] [CrossRef]
- Scheffer, D.d.L.; Latini, A. Exercise-Induced Immune System Response: Anti-Inflammatory Status on Peripheral and Central Organs. Biochim. Biophys. Acta BBA—Mol. Basis Dis. 2020, 1866, 165823. [Google Scholar] [CrossRef]
- Mathot, E.; Liberman, K.; Cao Dinh, H.; Njemini, R.; Bautmans, I. Systematic Review on the Effects of Physical Exercise on Cellular Immunosenescence-Related Markers—An Update. Exp. Gerontol. 2021, 149, 111318. [Google Scholar] [CrossRef]
- Landi, F.; Marzetti, E.; Martone, A.M.; Bernabei, R.; Onder, G. Exercise as a Remedy for Sarcopenia. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 25–31. [Google Scholar] [CrossRef]
- Gomes-Neto, M.; Saquetto, M.B.; Silva, C.M.; Carvalho, V.O.; Ribeiro, N.; Conceição, C.S. Effects of Respiratory Muscle Training on Respiratory Function, Respiratory Muscle Strength, and Exercise Tolerance in Patients Poststroke: A Systematic Review with Meta-Analysis. Arch. Phys. Med. Rehabil. 2016, 97, 1994–2001. [Google Scholar] [CrossRef] [PubMed]
- Stanford, K.I.; Goodyear, L.J. Exercise Regulation of Adipose Tissue. Adipocyte 2016, 5, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiuza-Luces, C.; Santos-Lozano, A.; Joyner, M.; Carrera-Bastos, P.; Picazo, O.; Zugaza, J.L.; Izquierdo, M.; Ruilope, L.M.; Lucia, A. Exercise Benefits in Cardiovascular Disease: Beyond Attenuation of Traditional Risk Factors. Nat. Rev. Cardiol. 2018, 15, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Bouassida, A.; Chamari, K.; Zaouali, M.; Feki, Y.; Zbidi, A.; Tabka, Z. Review on Leptin and Adiponectin Responses and Adaptations to Acute and Chronic Exercise. Br. J. Sports Med. 2010, 44, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Filgueira, T.O.; Castoldi, A.; Santos, L.E.R.; de Amorim, G.J.; de Sousa Fernandes, M.S.; Anastácio, W.d.L.d.N.; Campos, E.Z.; Santos, T.M.; Souto, F.O. The Relevance of a Physical Active Lifestyle and Physical Fitness on Immune Defense: Mitigating Disease Burden, with Focus on COVID-19 Consequences. Front. Immunol. 2021, 12, 587146. [Google Scholar] [CrossRef] [PubMed]
- Rehm, J.; Samokhvalov, A.V.; Neuman, M.G.; Room, R.; Parry, C.; Lönnroth, K.; Patra, J.; Poznyak, V.; Popova, S. The Association between Alcohol Use, Alcohol Use Disorders and Tuberculosis (TB). A Systematic Review. BMC Public Health 2009, 9, 450. [Google Scholar] [CrossRef]
- Simou, E.; Leonardi-Bee, J.; Britton, J. The Effect of Alcohol Consumption on the Risk of ARDS: A Systematic Review and Meta-Analysis. Chest 2018, 154, 58–68. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Gao, Y.; Shi, S.; Chen, Y.; Yang, K.; Tian, J. Drinking No-Links to the Severity of COVID-19: A Systematic Review and Meta-Analysis. J. Infect. 2020, 81, e126–e127. [Google Scholar] [CrossRef]
- Hamer, M.; Kivimäki, M.; Gale, C.R.; Batty, G.D. Lifestyle Risk Factors, Inflammatory Mechanisms, and COVID-19 Hospitalization: A Community-Based Cohort Study of 387,109 Adults in UK. Brain. Behav. Immun. 2020, 87, 184–187. [Google Scholar] [CrossRef]
- Dai, M.; Tao, L.; Chen, Z.; Tian, Z.; Guo, X.; Allen-Gipson, D.S.; Tan, R.; Li, R.; Chai, L.; Ai, F.; et al. Influence of Cigarettes and Alcohol on the Severity and Death of COVID-19: A Multicenter Retrospective Study in Wuhan, China. Front. Physiol. 2020, 11, 588553. [Google Scholar] [CrossRef]
- Bhalla, S.; Sharma, B.; Smith, D.; Boley, R.; McCluskey, C.; Ilyas, Y.; Afshar, M.; Balk, R.; Karnik, N.; Keshavarzian, A. Investigating Unhealthy Alcohol Use as an Independent Risk Factor for Increased COVID-19 Disease Severity: Observational Cross-Sectional Study. JMIR Public Health Surveill. 2021, 7, e33022. [Google Scholar] [CrossRef] [PubMed]
- Lassen, M.C.H.; Skaarup, K.G.; Sengeløv, M.; Iversen, K.; Ulrik, C.S.; Jensen, J.U.S.; Biering-Sørensen, T. Alcohol Consumption and the Risk of Acute Respiratory Distress Syndrome in COVID-19. Ann. Am. Thorac. Soc. 2021, 18, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Alberca, R.W.; Rigato, P.O.; Ramos, Y.Á.L.; Teixeira, F.M.E.; Branco, A.C.C.; Fernandes, I.G.; Pietrobon, A.J.; Duarte, A.J.d.S.; Aoki, V.; Orfali, R.L.; et al. Clinical Characteristics and Survival Analysis in Frequent Alcohol Consumers with COVID-19. Front. Nutr. 2021, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Wendt, F.R.; De Lillo, A.; Pathak, G.A.; De Angelis, F.; Polimanti, R. Host Genetic Liability for Severe COVID-19 Associates with Alcohol Drinking Behavior and Diabetic Outcomes in Participants of European Descent. Front. Genet. 2021, 12, 765247. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhou, H.; Hodgkinson, C.; Montero, A.; Goldman, D.; Chang, S.L. Network Meta-Analysis on the Mechanisms Underlying Alcohol Augmentation of COVID-19 Pathologies. Alcohol. Clin. Exp. Res. 2021, 45, 675–688. [Google Scholar] [CrossRef]
- Sureshchandra, S.; Raus, A.; Jankeel, A.; Ligh, B.J.K.; Walter, N.A.R.; Newman, N.; Grant, K.A.; Messaoudi, I. Dose-Dependent Effects of Chronic Alcohol Drinking on Peripheral Immune Responses. Sci. Rep. 2019, 9, 7847. [Google Scholar] [CrossRef]
- Yeligar, S.M.; Chen, M.M.; Kovacs, E.J.; Sisson, J.H.; Burnham, E.L.; Brown, L.A.S. Alcohol and Lung Injury and Immunity. Alcohol 2016, 55, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Ben-Eliyahu, S.; Page, G.G.; Yirmiya, R.; Taylor, A.N. Acute Alcohol Intoxication Suppresses Natural Killer Cell Activity and Promotes Tumor Metastasis. Nat. Med. 1996, 2, 457–460. [Google Scholar] [CrossRef]
- Sisson, J.H. Alcohol and Airways Function in Health and Disease. Alcohol 2007, 41, 293–307. [Google Scholar] [CrossRef] [Green Version]
- Arcavi, L.; Benowitz, N.L. Cigarette Smoking and Infection. Arch. Intern. Med. 2004, 164, 2206–2216. [Google Scholar] [CrossRef]
- Lacedonia, D.; Scioscia, G.; Santomasi, C.; Fuso, P.; Carpagnano, G.E.; Portacci, A.; Mastroianni, F.; Larizza, G.; Sabato, E.; Profilo, E.; et al. Impact of Smoking, COPD and Comorbidities on the Mortality of COVID-19 Patients. Sci. Rep. 2021, 11, 19251. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Baranova, A.; Cao, H.; Chen, J.; Zhang, X.; Zhang, F. Genetic Mechanisms of COVID-19 and Its Association with Smoking and Alcohol Consumption. Brief. Bioinform. 2021, 22, bbab284. [Google Scholar] [CrossRef] [PubMed]
- Clift, A.K.; von Ende, A.; Tan, P.S.; Sallis, H.M.; Lindson, N.; Coupland, C.A.C.; Munafò, M.R.; Aveyard, P.; Hippisley-Cox, J.; Hopewell, J.C. Smoking and COVID-19 Outcomes: An Observational and Mendelian Randomisation Study Using the UK Biobank Cohort. Thorax 2022, 77, 65–73. [Google Scholar] [CrossRef]
- Young-Wolff, K.C.; Slama, N.; Alexeeff, S.E.; Sakoda, L.C.; Fogelberg, R.; Myers, L.C.; Campbell, C.I.; Adams, A.S.; Prochaska, J.J. Tobacco Smoking and Risk of SARS-CoV-2 Infection and Disease Severity among Adults in an Integrated Healthcare System in California. Nicotine Tob. Res. 2023, 25, 211–220. [Google Scholar] [CrossRef]
- Puebla Neira, D.; Watts, A.; Seashore, J.; Polychronopoulou, E.; Kuo, Y.-F.; Sharma, G. Smoking and Risk of COVID-19 Hospitalization. Respir. Med. 2021, 182, 106414. [Google Scholar] [CrossRef] [PubMed]
- Simons, D.; Shahab, L.; Brown, J.; Perski, O. The Association of Smoking Status with SARS-CoV-2 Infection, Hospitalization and Mortality from COVID-19: A Living Rapid Evidence Review with Bayesian Meta-Analyses (Version 7). Addiction 2021, 116, 1319–1368. [Google Scholar] [CrossRef] [PubMed]
- Piasecki, T.M.; Smith, S.S.; Baker, T.B.; Slutske, W.S.; Adsit, R.T.; Bolt, D.M.; Conner, K.L.; Bernstein, S.L.; Eng, O.D.; Lazuk, D.; et al. Smoking Status, Nicotine Medication, Vaccination, and COVID-19 Hospital Outcomes: Findings from the COVID EHR Cohort at the University of Wisconsin (CEC-UW) Study. Nicotine Tob. Res. 2022, ntac201. [Google Scholar] [CrossRef]
- Hou, H.; Li, Y.; Zhang, P.; Wu, J.; Shi, L.; Xu, J.; Diao, J.; Wang, Y.; Yang, H. Smoking Is Independently Associated with an Increased Risk for COVID-19 Mortality: A Systematic Review and Meta-Analysis Based on Adjusted Effect Estimates. Nicotine Tob. Res. 2021, 23, 1947–1951. [Google Scholar] [CrossRef]
- Reddy, R.K.; Charles, W.N.; Sklavounos, A.; Dutt, A.; Seed, P.T.; Khajuria, A. The Effect of Smoking on COVID-19 Severity: A Systematic Review and Meta-Analysis. J. Med. Virol. 2021, 93, 1045–1056. [Google Scholar] [CrossRef]
- Patanavanich, R.; Glantz, S.A. Smoking Is Associated with Worse Outcomes of COVID-19 Particularly among Younger Adults: A Systematic Review and Meta-Analysis. BMC Public Health 2021, 21, 1554. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.; Daniels, L.B.; DeFilippis, A.P.; Hamburg, N.M.; Khan, Y.; Keith, R.J.; Kumar, R.S.; Strokes, A.C.; Robertson, R.M.; Bhatnagar, A. Smoking Is Associated with Increased Risk of Cardiovascular Events, Disease Severity, and Mortality among Patients Hospitalized for SARS-CoV-2 Infections. PLoS ONE 2022, 17, e0270763. [Google Scholar] [CrossRef] [PubMed]
- Farsalinos, K.; Barbouni, A.; Poulas, K.; Polosa, R.; Caponnetto, P.; Niaura, R. Current Smoking, Former Smoking, and Adverse Outcome among Hospitalized COVID-19 Patients: A Systematic Review and Meta-Analysis. Ther. Adv. Chronic Dis. 2020, 11, 2040622320935765. [Google Scholar] [CrossRef] [PubMed]
- Umnuaypornlert, A.; Kanchanasurakit, S.; Lucero-Prisno, D.E.I.; Saokaew, S. Smoking and Risk of Negative Outcomes among COVID-19 Patients: A Systematic Review and Meta-Analysis. Tob. Induc. Dis. 2021, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Saadatian-Elahi, M.; Amour, S.; Elias, C.; Henaff, L.; Dananché, C.; Vanhems, P. Tobacco Smoking and Severity of COVID-19: Experience from a Hospital-Based Prospective Cohort Study in Lyon, France. J. Med. Virol. 2021, 93, 6822–6827. [Google Scholar] [CrossRef]
- Benowitz, N.L.; Goniewicz, M.L.; Halpern-Felsher, B.; Krishnan-Sarin, S.; Ling, P.M.; O’Connor, R.J.; Pentz, M.A.; Robertson, R.M.; Bhatnagar, A. Tobacco Product Use and the Risks of SARS-CoV-2 Infection and COVID-19: Current Understanding and Recommendations for Future Research. Lancet Respir. Med. 2022, 10, 900–915. [Google Scholar] [CrossRef]
- Assad, N.A.; Kapoor, V.; Sood, A. Biomass Smoke Exposure and Chronic Lung Disease. Curr. Opin. Pulm. Med. 2016, 22, 150–157. [Google Scholar] [CrossRef]
- Kondo, T.; Nakano, Y.; Adachi, S.; Murohara, T. Effects of Tobacco Smoking on Cardiovascular Disease. Circ. J. 2019, 83, 1980–1985. [Google Scholar] [CrossRef] [Green Version]
- Zhu, P.; Pan, X.-F.; Sheng, L.; Chen, H.; Pan, A. Cigarette Smoking, Diabetes, and Diabetes Complications: Call for Urgent Action. Curr. Diab. Rep. 2017, 17, 78. [Google Scholar] [CrossRef]
- Habesoglu, M.; Demir, K.; Yumusakhuylu, A.C.; Sahin Yilmaz, A.; Oysu, C. Does Passive Smoking Have an Effect on Nasal Mucociliary Clearance? Otolaryngol. Neck Surg. 2012, 147, 152–156. [Google Scholar] [CrossRef]
- Morrison, D.; Rahman, I.; Lannan, S.; MacNEE, W. Epithelial Permeability, Inflammation, and Oxidant Stress in the Air Spaces of Smokers. Am. J. Respir. Crit. Care Med. 1999, 159, 473–479. [Google Scholar] [CrossRef]
- Johnson, J.D.; Houchens, D.P.; Kluwe, W.M.; Craig, D.K.; Fisher, G.L. Effects of Mainstream and Environmental Tobacco Smoke on the Immune System in Animals and Humans: A Review. Crit. Rev. Toxicol. 1990, 20, 369–395. [Google Scholar] [CrossRef] [PubMed]
- Purkayastha, A.; Sen, C.; Garcia, G.; Langerman, J.; Shia, D.W.; Meneses, L.K.; Vijayaraj, P.; Durra, A.; Koloff, C.R.; Freund, D.R.; et al. Direct Exposure to SARS-CoV-2 and Cigarette Smoke Increases Infection Severity and Alters the Stem Cell-Derived Airway Repair Response. Cell Stem Cell 2020, 27, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Burke, A.; FitzGerald, G.A. Oxidative Stress and Smoking-Induced Vascular Injury. Prog. Cardiovasc. Dis. 2003, 46, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, R.B.; Coletta, R.D.; Silvério, K.G.; Benevides, L.; Casati, M.Z.; da Silva, J.S.; Nociti, F.H. Impact of Smoking on Inflammation: Overview of Molecular Mechanisms. Inflamm. Res. 2011, 60, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xin, J.; Cai, S.; Jiang, X. Mendelian Randomization Analysis Provides Causality of Smoking on the Expression of ACE2, a Putative SARS-CoV-2 Receptor. eLife 2021, 10, e64188. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.C.; Sausville, E.L.; Girish, V.; Yuan, M.L.; Vasudevan, A.; John, K.M.; Sheltzer, J.M. Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract. Dev. Cell 2020, 53, 514–529.e3. [Google Scholar] [CrossRef]
- Akerström, S.; Mousavi-Jazi, M.; Klingström, J.; Leijon, M.; Lundkvist, A.; Mirazimi, A. Nitric Oxide Inhibits the Replication Cycle of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2005, 79, 1966–1969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Yu, M.; Ochani, M.; Amella, C.A.; Tanovic, M.; Susarla, S.; Li, J.H.; Wang, H.; Yang, H.; Ulloa, L.; et al. Nicotinic Acetylcholine Receptor Alpha7 Subunit Is an Essential Regulator of Inflammation. Nature 2003, 421, 384–388. [Google Scholar] [CrossRef]
- Pavlov, V.A.; Tracey, K.J. The Cholinergic Anti-Inflammatory Pathway. Brain. Behav. Immun. 2005, 19, 493–499. [Google Scholar] [CrossRef]
- Wells, J.C.; Sawaya, A.L.; Wibaek, R.; Mwangome, M.; Poullas, M.S.; Yajnik, C.S.; Demaio, A. The Double Burden of Malnutrition: Aetiological Pathways and Consequences for Health. The Lancet 2020, 395, 75–88. [Google Scholar] [CrossRef]
- Foolchand, A.; Ghazi, T.; Chuturgoon, A.A. Malnutrition and Dietary Habits Alter the Immune System Which May Consequently Influence SARS-CoV-2 Virulence: A Review. Int. J. Mol. Sci. 2022, 23, 2654. [Google Scholar] [CrossRef] [PubMed]
- Söderström, L.; Rosenblad, A.; Adolfsson, E.T.; Bergkvist, L. Malnutrition Is Associated with Increased Mortality in Older Adults Regardless of the Cause of Death. Br. J. Nutr. 2017, 117, 532–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponce, J.; Anzalone, A.J.; Bailey, K.; Sayles, H.; Timmerman, M.; Jackson, M.; McClay, J.; Hanson, C.; National COVID Cohort Collaborative (N3C) Consortium. Impact of Malnutrition on Clinical Outcomes in Patients Diagnosed with COVID-19. J. Parenter. Enter. Nutr. 2022, 46, 1797–1807. [Google Scholar] [CrossRef]
- Kurtz, A.; Grant, K.; Marano, R.; Arrieta, A.; Grant, K.; Feaster, W.; Steele, C.; Ehwerhemuepha, L. Long-Term Effects of Malnutrition on Severity of COVID-19. Sci. Rep. 2021, 11, 14974. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, Y.; Ge, Y.; Shi, Y.; Lv, P.; Zhang, J.; Fu, G.; Zhou, Y.; Jiang, K.; Lin, N.; et al. Evaluation of Nutrition Risk and Its Association with Mortality Risk in Severely and Critically Ill COVID-19 Patients. J. Parenter. Enter. Nutr. 2021, 45, 32–42. [Google Scholar] [CrossRef]
- Yu, Y.; Ye, J.; Chen, M.; Jiang, C.; Lin, W.; Lu, Y.; Ye, H.; Li, Y.; Wang, Y.; Liao, Q.; et al. Malnutrition Prolongs the Hospitalization of Patients with COVID-19 Infection: A Clinical Epidemiological Analysis. J. Nutr. Health Aging 2021, 25, 369–373. [Google Scholar] [CrossRef]
- Yue, Y.; Ma, W.; Accorsi, E.K.; Ding, M.; Hu, F.; Willett, W.C.; Chan, A.T.; Sun, Q.; Edwards, J.R.; Smith-Warner, S.A.; et al. Long-Term Diet and Risk of SARS-CoV-2 Infection and Coronavirus Disease 2019 (COVID-19) Severity. Am. J. Clin. Nutr. 2022, 116, 1672–1681. [Google Scholar] [CrossRef]
- Hou, Y.-C.; Su, W.-L.; Chao, Y.-C. COVID-19 Illness Severity in the Elderly in Relation to Vegetarian and Non-Vegetarian Diets: A Single-Center Experience. Front. Nutr. 2022, 9, 837458. [Google Scholar] [CrossRef]
- Kim, H.; Rebholz, C.M.; Hegde, S.; LaFiura, C.; Raghavan, M.; Lloyd, J.F.; Cheng, S.; Seidelmann, S.B. Plant-Based Diets, Pescatarian Diets and COVID-19 Severity: A Population-Based Case–Control Study in Six Countries. BMJ Nutr. Prev. Health 2021, 4, 257–266. [Google Scholar] [CrossRef]
- Merino, J.; Joshi, A.D.; Nguyen, L.H.; Leeming, E.R.; Mazidi, M.; Drew, D.A.; Gibson, R.; Graham, M.S.; Lo, C.-H.; Capdevila, J.; et al. Diet Quality and Risk and Severity of COVID-19: A Prospective Cohort Study. Gut 2021, 70, 2096–2104. [Google Scholar] [CrossRef]
- Trujillo-Mayol, I.; Guerra-Valle, M.; Casas-Forero, N.; Sobral, M.M.C.; Viegas, O.; Alarcón-Enos, J.; Ferreira, I.M.; Pinho, O. Western Dietary Pattern Antioxidant Intakes and Oxidative Stress: Importance During the SARS-CoV-2/COVID-19 Pandemic. Adv. Nutr. 2021, 12, 670–681. [Google Scholar] [CrossRef]
- Chrysohoou, C.; Panagiotakos, D.B.; Pitsavos, C.; Das, U.N.; Stefanadis, C. Adherence to the Mediterranean Diet Attenuates Inflammation and Coagulation Process in Healthy Adults. J. Am. Coll. Cardiol. 2004, 44, 152–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponzo, V.; Pellegrini, M.; D’Eusebio, C.; Bioletto, F.; Goitre, I.; Buscemi, S.; Frea, S.; Ghigo, E.; Bo, S. Mediterranean Diet and SARS-COV-2 Infection: Is There Any Association? A Proof-of-Concept Study. Nutrients 2021, 13, 1721. [Google Scholar] [CrossRef] [PubMed]
- Perez-Araluce, R.; Martinez-Gonzalez, M.A.; Fernández-Lázaro, C.I.; Bes-Rastrollo, M.; Gea, A.; Carlos, S. Mediterranean Diet and the Risk of COVID-19 in the ‘Seguimiento Universidad de Navarra’ Cohort. Clin. Nutr. 2021, 41, 3061–3068. [Google Scholar] [CrossRef] [PubMed]
- Horne, B.D.; Muhlestein, J.B.; May, H.T.; Le, V.T.; Bair, T.L.; Knowlton, K.U.; Anderson, J.L.; INSPIRE Registry Investigators. Association of Periodic Fasting with Lower Severity of COVID-19 Outcomes in the SARS-CoV-2 Pre-Vaccine Era: An Observational Cohort from the INSPIRE Registry. medRxiv 2022. [Google Scholar] [CrossRef]
- Sukkar, S.G.; Cogorno, L.; Pisciotta, L.; Pasta, A.; Vena, A.; Gradaschi, R.; Dentone, C.; Guiddo, E.; Martino, E.; Beltramini, S.; et al. Clinical Efficacy of Eucaloric Ketogenic Nutrition in the COVID-19 Cytokine Storm: A Retrospective Analysis of Mortality and Intensive Care Unit Admission. Nutrition 2021, 89, 111236. [Google Scholar] [CrossRef]
- Gangitano, E.; Tozzi, R.; Gandini, O.; Watanabe, M.; Basciani, S.; Mariani, S.; Lenzi, A.; Gnessi, L.; Lubrano, C. Ketogenic Diet as a Preventive and Supportive Care for COVID-19 Patients. Nutrients 2021, 13, 1004. [Google Scholar] [CrossRef]
- Ling, V.; Zabetakis, I. The Role of an Anti-Inflammatory Diet in Conjunction to COVID-19. Diseases 2021, 9, 76. [Google Scholar] [CrossRef]
- Julkunen, H.; Cichońska, A.; Slagboom, P.E.; Würtz, P. Nightingale Health UK Biobank Initiative Metabolic Biomarker Profiling for Identification of Susceptibility to Severe Pneumonia and COVID-19 in the General Population. eLife 2021, 10, e63033. [Google Scholar] [CrossRef]
- Zapata, B.R.; Müller, J.M.; Vásquez, J.E.; Ravera, F.; Lago, G.; Cañón, E.; Castañeda, D.; Pradenas, M.; Ramírez-Santana, M. Omega-3 Index and Clinical Outcomes of Severe COVID-19: Preliminary Results of a Cross-Sectional Study. Int. J. Environ. Res. Public. Health 2021, 18, 7722. [Google Scholar] [CrossRef]
- Mazidimoradi, A.; Alemzadeh, E.; Alemzadeh, E.; Salehiniya, H. The Effect of Polyunsaturated Fatty Acids on the Severity and Mortality of COVID Patients: A Systematic Review. Life Sci. 2022, 299, 120489. [Google Scholar] [CrossRef]
- Bejan, C.A.; Cahill, K.N.; Staso, P.J.; Choi, L.; Peterson, J.F.; Phillips, E.J. DrugWAS: Drug-Wide Association Studies for COVID-19 Drug Repurposing. Clin. Pharmacol. Ther. 2021, 110, 1537–1546. [Google Scholar] [CrossRef]
- Sedighiyan, M.; Abdollahi, H.; Karimi, E.; Badeli, M.; Erfanian, R.; Raeesi, S.; Hashemi, R.; Vahabi, Z.; Asanjarani, B.; Mansouri, F.; et al. Omega-3 Polyunsaturated Fatty Acids Supplementation Improve Clinical Symptoms in Patients with COVID-19: A Randomised Clinical Trial. Int. J. Clin. Pract. 2021, 75, e14854. [Google Scholar] [CrossRef] [PubMed]
- Langlois, P.L.; D’Aragon, F.; Hardy, G.; Manzanares, W. Omega-3 Polyunsaturated Fatty Acids in Critically Ill Patients with Acute Respiratory Distress Syndrome: A Systematic Review and Meta-Analysis. Nutrition 2019, 61, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Chiang, N. Resolution Phase Lipid Mediators of Inflammation: Agonists of Resolution. Curr. Opin. Pharmacol. 2013, 13, 632–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serhan, C.N. Pro-Resolving Lipid Mediators Are Leads for Resolution Physiology. Nature 2014, 510, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Zaloga, G.P. Narrative Review of N-3 Polyunsaturated Fatty Acid Supplementation upon Immune Functions, Resolution Molecules and Lipid Peroxidation. Nutrients 2021, 13, 662. [Google Scholar] [CrossRef]
- Schwarz, B.; Sharma, L.; Roberts, L.; Peng, X.; Bermejo, S.; Leighton, I.; Casanovas-Massana, A.; Minasyan, M.; Farhadian, S.; Ko, A.I.; et al. Cutting Edge: Severe SARS-CoV-2 Infection in Humans Is Defined by a Shift in the Serum Lipidome, Resulting in Dysregulation of Eicosanoid Immune Mediators. J. Immunol. 2021, 206, 329–334. [Google Scholar] [CrossRef]
- Goc, A.; Niedzwiecki, A.; Rath, M. Polyunsaturated ω-3 Fatty Acids Inhibit ACE2-Controlled SARS-CoV-2 Binding and Cellular Entry. Sci. Rep. 2021, 11, 5207. [Google Scholar] [CrossRef]
- Brockman-Schneider, R.A.; Pickles, R.J.; Gern, J.E. Effects of Vitamin D on Airway Epithelial Cell Morphology and Rhinovirus Replication. PLoS ONE 2014, 9, e86755. [Google Scholar] [CrossRef] [Green Version]
- Hansdottir, S.; Monick, M.M.; Lovan, N.; Powers, L.; Gerke, A.; Hunninghake, G.W. Vitamin D Decreases Respiratory Syncytial Virus Induction of NF-ΚB–Linked Chemokines and Cytokines in Airway Epithelium While Maintaining the Antiviral State. J. Immunol. 2010, 184, 965–974. [Google Scholar] [CrossRef] [Green Version]
- Cannell, J.J.; Vieth, R.; Umhau, J.C.; Holick, M.F.; Grant, W.B.; Madronich, S.; Garland, C.F.; Giovannucci, E. Epidemic Influenza and Vitamin D. Epidemiol. Infect. 2006, 134, 1129–1140. [Google Scholar] [CrossRef]
- Martineau, A.R.; Cantorna, M.T. Vitamin D for COVID-19: Where Are We Now? Nat. Rev. Immunol. 2022, 22, 529–530. [Google Scholar] [CrossRef] [PubMed]
- Eggersdorfer, M.; Berger, M.M.; Calder, P.C.; Gombart, A.F.; Ho, E.; Laviano, A.; Meydani, S.N. Perspective: Role of Micronutrients and Omega-3 Long-Chain Polyunsaturated Fatty Acids for Immune Outcomes of Relevance to Infections in Older Adults—A Narrative Review and Call for Action. Adv. Nutr. 2022, 13, 1415–1430. [Google Scholar] [CrossRef] [PubMed]
- Orchard, L.; Baldry, M.; Nasim-Mohi, M.; Monck, C.; Saeed, K.; Grocott, M.P.W.; Ahilanandan, D. Vitamin-D Levels and Intensive Care Unit Outcomes of a Cohort of Critically Ill COVID-19 Patients. Clin. Chem. Lab. Med. CCLM 2021, 59, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Campi, I.; Gennari, L.; Merlotti, D.; Mingiano, C.; Frosali, A.; Giovanelli, L.; Torlasco, C.; Pengo, M.F.; Heilbron, F.; Soranna, D.; et al. Vitamin D and COVID-19 Severity and Related Mortality: A Prospective Study in Italy. BMC Infect. Dis. 2021, 21, 566. [Google Scholar] [CrossRef]
- Ramirez-Sandoval, J.C.; Castillos-Ávalos, V.J.; Paz-Cortés, A.; Santillan-Ceron, A.; Hernandez-Jimenez, S.; Mehta, R.; Correa-Rotter, R. Very Low Vitamin D Levels Are a Strong Independent Predictor of Mortality in Hospitalized Patients with Severe COVID-19. Arch. Med. Res. 2022, 53, 215–222. [Google Scholar] [CrossRef]
- Dissanayake, H.A.; de Silva, N.L.; Sumanatilleke, M.; de Silva, S.D.N.; Gamage, K.K.K.; Dematapitiya, C.; Kuruppu, D.C.; Ranasinghe, P.; Pathmanathan, S.; Katulanda, P. Prognostic and Therapeutic Role of Vitamin D in COVID-19: Systematic Review and Meta-Analysis. J. Clin. Endocrinol. Metab. 2022, 107, 1484–1502. [Google Scholar] [CrossRef]
- D’Ecclesiis, O.; Gavioli, C.; Martinoli, C.; Raimondi, S.; Chiocca, S.; Miccolo, C.; Bossi, P.; Cortinovis, D.; Chiaradonna, F.; Palorini, R.; et al. Vitamin D and SARS-CoV2 Infection, Severity and Mortality: A Systematic Review and Meta-Analysis. PLoS ONE 2022, 17, e0268396. [Google Scholar] [CrossRef]
- Loucera, C.; Peña-Chilet, M.; Esteban-Medina, M.; Muñoyerro-Muñiz, D.; Villegas, R.; Lopez-Miranda, J.; Rodriguez-Baño, J.; Túnez, I.; Bouillon, R.; Dopazo, J.; et al. Real World Evidence of Calcifediol or Vitamin D Prescription and Mortality Rate of COVID-19 in a Retrospective Cohort of Hospitalized Andalusian Patients. Sci. Rep. 2021, 11, 23380. [Google Scholar] [CrossRef]
- Nielsen, N.M.; Junker, T.G.; Cohen, A.S.; Munger, K.L.; Stenager, E.; Ascherio, A.; Boding, L.; Hviid, A. Vitamin D Status and Severity of COVID-19. Sci. Rep. 2022, 12, 19823. [Google Scholar] [CrossRef]
- Butler-Laporte, G.; Nakanishi, T.; Mooser, V.; Morrison, D.R.; Abdullah, T.; Adeleye, O.; Mamlouk, N.; Kimchi, N.; Afrasiabi, Z.; Rezk, N.; et al. Vitamin D and COVID-19 Susceptibility and Severity in the COVID-19 Host Genetics Initiative: A Mendelian Randomization Study. PLOS Med. 2021, 18, e1003605. [Google Scholar] [CrossRef]
- Freitas, A.T.; Calhau, C.; Antunes, G.; Araújo, B.; Bandeira, M.; Barreira, S.; Bazenga, F.; Braz, S.; Caldeira, D.; Santos, S.C.R.; et al. Vitamin D-Related Polymorphisms and Vitamin D Levels as Risk Biomarkers of COVID-19 Disease Severity. Sci. Rep. 2021, 11, 20837. [Google Scholar] [CrossRef] [PubMed]
- Hafezi, S.; Saheb Sharif-Askari, F.; Saheb Sharif-Askari, N.; Ali Hussain Alsayed, H.; Alsafar, H.; Al Anouti, F.; Hamid, Q.; Halwani, R. Vitamin D Enhances Type I IFN Signaling in COVID-19 Patients. Sci. Rep. 2022, 12, 17778. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.; Fraser, C.; Herbeck, J.T. A Strong Case for Viral Genetic Factors in HIV Virulence. Viruses 2011, 3, 204–216. [Google Scholar] [CrossRef]
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of Mortality in Patients Infected with SARS-CoV-2 Variant of Concern 202012/1: Matched Cohort Study. BMJ 2021, 372, n579. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Jarvis, C.I.; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef]
- Fisman, D.N.; Tuite, A.R. Evaluation of the Relative Virulence of Novel SARS-CoV-2 Variants: A Retrospective Cohort Study in Ontario, Canada. CMAJ 2021, 193, E1619–E1625. [Google Scholar] [CrossRef] [PubMed]
- Funk, T.; Pharris, A.; Spiteri, G.; Bundle, N.; Melidou, A.; Carr, M.; Gonzalez, G.; Garcia-Leon, A.; Crispie, F.; O’Connor, L.; et al. Characteristics of SARS-CoV-2 Variants of Concern B.1.1.7, B.1.351 or P.1: Data from Seven EU/EEA Countries, Weeks 38/2020 to 10/2021. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2021, 26, 2100348. [Google Scholar] [CrossRef] [PubMed]
- Abu-Raddad, L.J.; Chemaitelly, H.; Ayoub, H.H.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; Tang, P.; Hasan, M.R.; Coyle, P.; AlMukdad, S.; et al. Severity, Criticality, and Fatality of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Beta Variant. Clin. Infect. Dis. 2021, 75, e1188–e1191. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital Admission and Emergency Care Attendance Risk for SARS-CoV-2 Delta (B.1.617.2) Compared with Alpha (B.1.1.7) Variants of Concern: A Cohort Study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Sheikh, A.; McMenamin, J.; Taylor, B.; Robertson, C. Public Health Scotland and the EAVE II Collaborators SARS-CoV-2 Delta VOC in Scotland: Demographics, Risk of Hospital Admission, and Vaccine Effectiveness. Lancet Lond. Engl. 2021, 397, 2461–2462. [Google Scholar] [CrossRef]
- Peralta-Santos, A.; Rodrigues, E.F.; Moreno, J.; Ricoca, V.; Casaca, P.; Fernandes, E.; Gomes, J.P.; Ferreira, R.; Isidro, J.; Pinto, M.; et al. Omicron (BA.1) SARS-CoV-2 Variant Is Associated with Reduced Risk of Hospitalization and Length of Stay Compared with Delta (B.1.617.2). medRxiv 2022. [Google Scholar] [CrossRef]
- Lewnard, J.A.; Hong, V.X.; Patel, M.M.; Kahn, R.; Lipsitch, M.; Tartof, S.Y. Clinical Outcomes among Patients Infected with Omicron (B.1.1.529) SARS-CoV-2 Variant in Southern California. medRxiv 2022. [Google Scholar] [CrossRef]
- Davies, M.-A.; Kassanjee, R.; Rousseau, P.; Morden, E.; Johnson, L.; Solomon, W.; Hsiao, N.-Y.; Hussey, H.; Meintjes, G.; Paleker, M.; et al. Outcomes of Laboratory-Confirmed SARS-CoV-2 Infection in the Omicron-Driven Fourth Wave Compared with Previous Waves in the Western Cape Province, South Africa. Trop. Med. Int. Health 2022, 27, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Strasser, Z.H.; Greifer, N.; Hadavand, A.; Murphy, S.N.; Estiri, H. Estimates of SARS-CoV-2 Omicron BA.2 Subvariant Severity in New England. JAMA Netw. Open 2022, 5, e2238354. [Google Scholar] [CrossRef] [PubMed]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C.; et al. Early Assessment of the Clinical Severity of the SARS-CoV-2 Omicron Variant in South Africa: A Data Linkage Study. Lancet 2022, 399, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.E.; Dávila-Barclay, A.; Salvatierra, G.; González, L.; Cuicapuza, D.; Solís, L.; Marcos-Carbajal, P.; Huancachoque, J.; Maturrano, L.; Tsukayama, P. The Emergence of Sars-CoV-2 Variant Lambda (C.37) in South America. Microbiol. Spectr. 2021, 9, e00789-21. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, R.P.; Hanage, W.P. Challenges in Inferring Intrinsic Severity of the SARS-CoV-2 Omicron Variant. N. Engl. J. Med. 2022, 386, e14. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.-C.; Ng, K.-C.; Ching, R.H.H.; Lai, K.-L.; Kam, T.T.; Gu, H.; Sit, K.-Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron Variant Replication in Human Bronchus and Lung Ex Vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Abdelnabi, R.; Foo, C.S.; Zhang, X.; Lemmens, V.; Maes, P.; Slechten, B.; Raymenants, J.; André, E.; Weynand, B.; Dallemier, K.; et al. The Omicron (B.1.1.529) SARS-CoV-2 Variant of Concern Does Not Readily Infect Syrian Hamsters. Microbiology 2022, 198, 105253. [Google Scholar] [CrossRef]
- Chen, D.-Y.; Kenney, D.; Chin, C.V.; Tavares, A.H.; Khan, N.; Conway, H.L.; Liu, G.; Choudhary, M.C.; Gertje, H.P.; O’Connell, A.K.; et al. Role of Spike in the Pathogenic and Antigenic Behavior of SARS-CoV-2 BA.1 Omicron. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kudriavtsev, A.V.; Vakhrusheva, A.V.; Novoseletsky, V.N.; Bozdaganyan, M.E.; Shaitan, K.V.; Kirpichnikov, M.P.; Sokolova, O.S. Immune Escape Associated with RBD Omicron Mutations and SARS-CoV-2 Evolution Dynamics. Viruses 2022, 14, 1603. [Google Scholar] [CrossRef]
- Butt, A.A.; Dargham, S.R.; Coyle, P.; Yassine, H.M.; Al-Khal, A.; Abou-Samra, A.-B.; Abu-Raddad, L.J. COVID-19 Disease Severity in Persons Infected with Omicron BA.1 and BA.2 Sublineages and Association with Vaccination Status. JAMA Intern. Med. 2022, 182, 1097–1099. [Google Scholar] [CrossRef] [PubMed]
- Sievers, C.; Zacher, B.; Ullrich, A.; Huska, M.; Fuchs, S.; Buda, S.; Haas, W.; Diercke, M.; Heiden, M.A.D.; Kröger, S. SARS-CoV-2 Omicron Variants BA.1 and BA.2 Both Show Similarly Reduced Disease Severity of COVID-19 Compared to Delta, Germany, 2021 to 2022. Eurosurveillance 2022, 27, 2200396. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.-A.; Morden, E.; Rosseau, P.; Arendse, J.; Bam, J.-L.; Boloko, L.; Cloete, K.; Cohen, C.; Chetty, N.; Dane, P.; et al. Outcomes of Laboratory-Confirmed SARS-CoV-2 Infection during Resurgence Driven by Omicron Lineages BA.4 and BA.5 Compared with Previous Waves in the Western Cape Province, South Africa. Int. J. Infect. Dis. 2023, 127, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Lewnard, J.A.; Hong, V.; Tartof, S.Y. Association of SARS-CoV-2 BA.4/BA.5 Omicron Lineages with Immune Escape and Clinical Outcome. medRxiv 2022. [Google Scholar] [CrossRef]
- Hansen, C.H.; Friis, N.U.; Bager, P.; Stegger, M.; Fonager, J.; Fomsgaard, A.; Gram, M.A.; Christiansen, L.E.; Ethelberg, S.; Legarth, R.; et al. Risk of Reinfection, Vaccine Protection, and Severity of Infection with the BA.5 Omicron Subvariant: A Nation-Wide Population-Based Study in Denmark. Lancet Infect. Dis. 2022. [Google Scholar] [CrossRef]
- Markov, P.V.; Katzourakis, A.; Stilianakis, N.I. Antigenic Evolution Will Lead to New SARS-CoV-2 Variants with Unpredictable Severity. Nat. Rev. Microbiol. 2022, 20, 251–252. [Google Scholar] [CrossRef] [PubMed]
- Mourier, T.; Shuaib, M.; Hala, S.; Mfarrej, S.; Alofi, F.; Naeem, R.; Alsomali, A.; Jorgensen, D.; Subudhi, A.K.; Ben Rached, F.; et al. SARS-CoV-2 Genomes from Saudi Arabia Implicate Nucleocapsid Mutations in Host Response and Increased Viral Load. Nat. Commun. 2022, 13, 601. [Google Scholar] [CrossRef] [PubMed]
- Dumonteil, E.; Fusco, D.; Drouin, A.; Herrera, C. Genomic Signatures of SARS-CoV-2 Associated with Patient Mortality. Viruses 2021, 13, 227. [Google Scholar] [CrossRef]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef]
- Lee, A.C.-Y.; Zhang, A.J.; Chan, J.F.-W.; Li, C.; Fan, Z.; Liu, F.; Chen, Y.; Liang, R.; Sridhar, S.; Cai, J.-P.; et al. Oral SARS-CoV-2 Inoculation Establishes Subclinical Respiratory Infection with Virus Shedding in Golden Syrian Hamsters. Cell Rep. Med. 2020, 1, 100121. [Google Scholar] [CrossRef]
- Ryan, K.A.; Bewley, K.R.; Fotheringham, S.A.; Slack, G.S.; Brown, P.; Hall, Y.; Wand, N.I.; Marriott, A.C.; Cavell, B.E.; Tree, J.A.; et al. Dose-Dependent Response to Infection with SARS-CoV-2 in the Ferret Model and Evidence of Protective Immunity. Nat. Commun. 2021, 12, 81. [Google Scholar] [CrossRef]
- Imai, M.; Iwatsuki-Horimoto, K.; Hatta, M.; Loeber, S.; Halfmann, P.J.; Nakajima, N.; Watanabe, T.; Ujie, M.; Takahashi, K.; Ito, M.; et al. Syrian Hamsters as a Small Animal Model for SARS-CoV-2 Infection and Countermeasure Development. Proc. Natl. Acad. Sci. USA 2020, 117, 16587–16595. [Google Scholar] [CrossRef]
- Gandhi, M.; Beyrer, C.; Goosby, E. Masks Do More Than Protect Others During COVID-19: Reducing the Inoculum of SARS-CoV-2 to Protect the Wearer. J. Gen. Intern. Med. 2020, 35, 3063–3066. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, M.A.; Glidden, D.V.; Gennatas, E.D.; Bielecki, M.; Beyrer, C.; Rutherford, G.; Chambers, H.; Goosby, E.; Gandhi, M. Importance of Non-Pharmaceutical Interventions in Lowering the Viral Inoculum to Reduce Susceptibility to Infection by SARS-CoV-2 and Potentially Disease Severity. Lancet Infect. Dis. 2021, 21, e296–e301. [Google Scholar] [CrossRef] [PubMed]
- Guallar, M.P.; Meiriño, R.; Donat-Vargas, C.; Corral, O.; Jouvé, N.; Soriano, V. Inoculum at the Time of SARS-CoV-2 Exposure and Risk of Disease Severity. Int. J. Infect. Dis. 2020, 97, 290–292. [Google Scholar] [CrossRef] [PubMed]
- Bielecki, M.; Züst, R.; Siegrist, D.; Meyerhofer, D.; Crameri, G.A.G.; Stanga, Z.; Stettbacher, A.; Buehrer, T.W.; Deuel, J.W. Social Distancing Alters the Clinical Course of COVID-19 in Young Adults: A Comparative Cohort Study. Clin. Infect. Dis. 2021, 72, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, W.; Dahake, R.; van de Pas, R.; Vanham, G.; Assefa, Y. COVID-19: Does the Infectious Inoculum Dose-Response Relationship Contribute to Understanding Heterogeneity in Disease Severity and Transmission Dynamics? Med. Hypotheses 2021, 146, 110431. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Czajkowski, L.M.; Donaldson, A.; Baus, H.A.; Reed, S.M.; Athota, R.S.; Bristol, T.; Rosas, L.A.; Cervantes-Medina, A.; Taubenberger, J.K.; et al. A Dose-Finding Study of a Wild-Type Influenza A(H3N2) Virus in a Healthy Volunteer Human Challenge Model. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 69, 2082–2090. [Google Scholar] [CrossRef]
- Memoli, M.J.; Czajkowski, L.; Reed, S.; Athota, R.; Bristol, T.; Proudfoot, K.; Fargis, S.; Stein, M.; Dunfee, R.L.; Shaw, P.A.; et al. Validation of the Wild-Type Influenza A Human Challenge Model H1N1pdMIST: An A(H1N1)Pdm09 Dose-Finding Investigational New Drug Study. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2015, 60, 693–702. [Google Scholar] [CrossRef] [Green Version]
- Yu, I.T.S.; Li, Y.; Wong, T.W.; Tam, W.; Chan, A.T.; Lee, J.H.W.; Leung, D.Y.C.; Ho, T. Evidence of Airborne Transmission of the Severe Acute Respiratory Syndrome Virus. N. Engl. J. Med. 2004, 350, 1731–1739. [Google Scholar] [CrossRef] [Green Version]
- Mills, J.; Van Kirk, J.E.; Wright, P.F.; Chanock, R.M. Experimental Respiratory Syncytial Virus Infection of Adults. Possible Mechanisms of Resistance to Infection and Illness. J. Immunol. Baltim. Md 1950 1971, 107, 123–130. [Google Scholar]
- Little, P.; Read, R.C.; Amlôt, R.; Chadborn, T.; Rice, C.; Bostock, J.; Yardley, L. Reducing Risks from Coronavirus Transmission in the Home—The Role of Viral Load. BMJ 2020, 369, m1728. [Google Scholar] [CrossRef]
- Bixler, S.L.; Stefan, C.P.; Jay, A.N.; Rossi, F.D.; Ricks, K.M.; Shoemaker, C.J.; Moreau, A.M.; Zeng, X.; Hooper, J.W.; Dyer, D.N.; et al. Exposure Route Influences Disease Severity in the COVID-19 Cynomolgus Macaque Model. Viruses 2022, 14, 1013. [Google Scholar] [CrossRef] [PubMed]
- Port, J.R.; Yinda, C.K.; Owusu, I.O.; Holbrook, M.; Fischer, R.; Bushmaker, T.; Avanzato, V.A.; Schulz, J.E.; Martens, C.; van Doremalen, N.; et al. SARS-CoV-2 Disease Severity and Transmission Efficiency Is Increased for Airborne Compared to Fomite Exposure in Syrian Hamsters. Nat. Commun. 2021, 12, 4985. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, L.M.; Escandón, K.; Ulrich, A.K.; Rasmussen, A.L.; Roy, C.J.; Bix, G.J.; Popescu, S.V.; Moore, K.A.; Osterholm, M.T. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Dose, Infection, and Disease Outcomes for Coronavirus Disease 2019 (COVID-19): A Review. Clin. Infect. Dis. 2022, 75, e1195–e1201. [Google Scholar] [CrossRef] [PubMed]
- University of Oxford. A Dose Finding Human Experimental Infection Study with SARS-CoV-2 in Healthy Volunteers with Immunologically Sensitised with Either Previous, Microbiologically Confirmed, SARS-CoV-2 Infection and/or Vaccination against SARS-CoV2; University of Oxford: Oxford, UK, 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT04864548 (accessed on 11 December 2022).
- Bilal, U.; Cainzos-Achirica, M.; Cleries, M.; Santaeugènia, S.; Corbella, X.; Comin-Colet, J.; Vela, E. Socioeconomic Status, Life Expectancy and Mortality in a Universal Healthcare Setting: An Individual-Level Analysis of >6 Million Catalan Residents. Prev. Med. 2019, 123, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Sanzenbacher, G.T.; Webb, A.; Cosgrove, C.M.; Orlova, N. Rising Inequality in Life Expectancy by Socioeconomic Status. N. Am. Actuar. J. 2021, 25, S566–S581. [Google Scholar] [CrossRef] [Green Version]
- Alkire, S.; Dirksen, J.; Nogales, R.; Oldiges, C. Multidimensional Poverty and COVID-19 Risk Factors: A Rapid Overview of Interlinked Deprivations across 5.8 Billion People. In Oxford Poverty and Human Development Initiative (OPHI); University of Oxford: Oxford, UK, 2021; Volume 53a, pp. 1–12. [Google Scholar]
- Desmet, K.; Wacziarg, R. JUE Insight: Understanding Spatial Variation in COVID-19 across the United States. J. Urban Econ. 2022, 127, 103332. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, X.; Xie, J. Spatial Inequalities of COVID-19 Mortality Rate in Relation to Socioeconomic and Environmental Factors across England. Sci. Total Environ. 2021, 758, 143595. [Google Scholar] [CrossRef]
- Abedi, V.; Olulana, O.; Avula, V.; Chaudhary, D.; Khan, A.; Shahjouei, S.; Li, J.; Zand, R. Racial, Economic, and Health Inequality and COVID-19 Infection in the United States. J. Racial Ethn. Health Disparities 2021, 8, 732–742. [Google Scholar] [CrossRef]
- Wachtler, B.; Michalski, N.; Nowossadeck, E.; Diercke, M.; Wahrendorf, M.; Santos-Hövener, C.; Lampert, T.; Hoebel, J. Socioeconomic Inequalities and COVID-19—A Review of the Current International Literature. J. Health Monit. 2020, 5, 3–17. [Google Scholar] [CrossRef]
- Martín-Sánchez, F.J.; Valls Carbó, A.; Miró, Ò.; Llorens, P.; Jiménez, S.; Piñera, P.; Burillo-Putze, G.; Martín, A.; García-Lamberechts, J.E.; Jacob, J.; et al. Socio-Demographic Health Determinants Are Associated with Poor Prognosis in Spanish Patients Hospitalized with COVID-19. J. Gen. Intern. Med. 2021, 36, 3737–3742. [Google Scholar] [CrossRef]
- Vaughan, L.; Veruttipong, D.; Shaw, J.G.; Levy, N.; Edwards, L.; Winget, M. Relationship of Socio-Demographics, Comorbidities, Symptoms and Healthcare Access with Early COVID-19 Presentation and Disease Severity. BMC Infect. Dis. 2021, 21, 40. [Google Scholar] [CrossRef] [PubMed]
- Ingraham, N.E.; Purcell, L.N.; Karam, B.S.; Dudley, R.A.; Usher, M.G.; Warlick, C.A.; Allen, M.L.; Melton, G.B.; Charles, A.; Tignanelli, C.J. Racial and Ethnic Disparities in Hospital Admissions from COVID-19: Determining the Impact of Neighborhood Deprivation and Primary Language. J. Gen. Intern. Med. 2021, 36, 3462–3470. [Google Scholar] [CrossRef] [PubMed]
- Little, C.; Alsen, M.; Barlow, J.; Naymagon, L.; Tremblay, D.; Genden, E.; Trosman, S.; Iavicoli, L.; van Gerwen, M. The Impact of Socioeconomic Status on the Clinical Outcomes of COVID-19; a Retrospective Cohort Study. J. Community Health 2021, 46, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Montoya, J.; Ballesteros, S.M.; Idrovo, A.J. COVID-19 Distribution in Bogotá, Colombia: Effect of Poverty during the First 2 Months of Pandemic. J. Epidemiol. Community Health 2022, 76, 116–120. [Google Scholar] [CrossRef]
- Broad, J.; Forman, J.; Brighouse, J.; Sobande, A.; McIntosh, A.; Watterson, C.; Boot, E.; Montgomery, F.; Gilmour, I.; Tan, J.; et al. Post-COVID-19 Paediatric Inflammatory Multisystem Syndrome: Association of Ethnicity, Key Worker and Socioeconomic Status with Risk and Severity. Arch. Dis. Child. 2021, 106, 1218–1225. [Google Scholar] [CrossRef]
- Ríos, V.; Denova-Gutiérrez, E.; Barquera, S. Association between Living in Municipalities with High Crowding Conditions and Poverty and Mortality from COVID-19 in Mexico. PLoS ONE 2022, 17, e0264137. [Google Scholar] [CrossRef]
- Millán-Guerrero, R.O.; Caballero-Hoyos, R.; Monárrez-Espino, J. Poverty and Survival from COVID-19 in Mexico. J. Public Health 2021, 43, 437–444. [Google Scholar] [CrossRef]
- Sesé, L.; Nguyen, Y.; Leprieur, E.G.; Annesi-Maesano, I.; Cavalin, C.; Bouillé, J.G.d.; Demestier, L.; Dhote, R.; Tandjaoui-Lambiotte, Y.; Bauvois, A.; et al. Impact of Socioeconomic Status in Patients Hospitalised for COVID-19 in the Greater Paris Area. Eur. Respir. J. 2020, 56, 2002364. [Google Scholar] [CrossRef]
- Lone, N.I.; McPeake, J.; Stewart, N.I.; Blayney, M.C.; Seem, R.C.; Donaldson, L.; Glass, E.; Haddow, C.; Hall, R.; Martin, C.; et al. Influence of Socioeconomic Deprivation on Interventions and Outcomes for Patients Admitted with COVID-19 to Critical Care Units in Scotland: A National Cohort Study. Lancet Reg. Health—Eur. 2021, 1, 100005. [Google Scholar] [CrossRef]
- Chung, G.K.-K.; Chan, S.-M.; Chan, Y.-H.; Yip, T.C.-F.; Ma, H.-M.; Wong, G.L.-H.; Chung, R.Y.-N.; Wong, H.; Wong, S.Y.-S.; Yeoh, E.K.; et al. Differential Impacts of Multimorbidity on COVID-19 Severity across the Socioeconomic Ladder in Hong Kong: A Syndemic Perspective. Int. J. Environ. Res. Public. Health 2021, 18, 8168. [Google Scholar] [CrossRef]
- Zakeri, R.; Bendayan, R.; Ashworth, M.; Bean, D.M.; Dodhia, H.; Durbaba, S.; O’Gallagher, K.; Palmer, C.; Curcin, V.; Aitken, E.; et al. A Case-Control and Cohort Study to Determine the Relationship between Ethnic Background and Severe COVID-19. EClinicalMedicine 2020, 28, 100574. [Google Scholar] [CrossRef]
- Patel, A.P.; Paranjpe, M.D.; Kathiresan, N.P.; Rivas, M.A.; Khera, A.V. Race, Socioeconomic Deprivation, and Hospitalization for COVID-19 in English Participants of a National Biobank. Int. J. Equity Health 2020, 19, 114. [Google Scholar] [CrossRef]
- Razieh, C.; Zaccardi, F.; Islam, N.; Gillies, C.L.; Chudasama, Y.V.; Rowlands, A.; Kloecker, D.E.; Davies, M.J.; Khunti, K.; Yates, T. Ethnic Minorities and COVID-19: Examining Whether Excess Risk Is Mediated through Deprivation. Eur. J. Public Health 2021, 31, 630–634. [Google Scholar] [CrossRef]
- Agyemang, C.; Richters, A.; Jolani, S.; Hendriks, S.; Zalpuri, S.; Yu, E.; Pijls, B.; Prins, M.; Stronks, K.; Zeegers, M.P. Ethnic Minority Status as Social Determinant for COVID-19 Infection, Hospitalisation, Severity, ICU Admission and Deaths in the Early Phase of the Pandemic: A Meta-Analysis. BMJ Glob. Health 2021, 6, e007433. [Google Scholar] [CrossRef]
- Mackey, K.; Ayers, C.K.; Kondo, K.K.; Saha, S.; Advani, S.M.; Young, S.; Spencer, H.; Rusek, M.; Anderson, J.; Veazie, S.; et al. Racial and Ethnic Disparities in COVID-19–Related Infections, Hospitalizations, and Deaths: A Systematic Review. Ann. Intern. Med. 2021, 174, 362–373. [Google Scholar] [CrossRef]
- Arasteh, K. Prevalence of Comorbidities and Risks Associated with COVID-19 among Black and Hispanic Populations in New York City: An Examination of the 2018 New York City Community Health Survey. J. Racial Ethn. Health Disparities 2021, 8, 863–869. [Google Scholar] [CrossRef]
- Lee, H.; Shin, S.H.; Gu, S.; Zhao, D.; Kang, D.; Joi, Y.R.; Suh, G.Y.; Pastor-Barriuso, R.; Guallar, E.; Cho, J.; et al. Racial Differences in Comorbidity Profile among Patients with Chronic Obstructive Pulmonary Disease. BMC Med. 2018, 16, 178. [Google Scholar] [CrossRef] [Green Version]
- Daw, J. Contribution of Four Comorbid Conditions to Racial/Ethnic Disparities in Mortality Risk. Am. J. Prev. Med. 2017, 52, S95–S102. [Google Scholar] [CrossRef]
- Cossrow, N.; Falkner, B. Race/Ethnic Issues in Obesity and Obesity-Related Comorbidities. J. Clin. Endocrinol. Metab. 2004, 89, 2590–2594. [Google Scholar] [CrossRef] [Green Version]
- Paeratakul, S.; Lovejoy, J.C.; Ryan, D.H.; Bray, G.A. The Relation of Gender, Race and Socioeconomic Status to Obesity and Obesity Comorbidities in a Sample of US Adults. Int. J. Obes. 2002, 26, 1205–1210. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.R. Race/Ethnicity and Socioeconomic Status: Measurement and Methodological Issues. Int. J. Health Serv. 1996, 26, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.; Brimblecombe, P.; Burrows, J.; Heal, M.R.; Grennfelt, P.; Stevenson, D.S.; Jowett, A.; Nemitz, E.; Coyle, M.; Liu, X.; et al. A Chronology of Global Air Quality. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2020, 378, 20190314. [Google Scholar] [CrossRef]
- Rzymski, P.; Poniedziałek, B.; Rosińska, J.; Rogalska, M.; Zarębska-Michaluk, D.; Rorat, M.; Moniuszko-Malinowska, A.; Lorenc, B.; Kozielewicz, D.; Piekarska, A.; et al. The Association of Airborne Particulate Matter and Benzo[a]Pyrene with the Clinical Course of COVID-19 in Patients Hospitalized in Poland. Environ. Pollut. 2022, 306, 119469. [Google Scholar] [CrossRef] [PubMed]
- Kogevinas, M.; Castaño-Vinyals, G.; Karachaliou, M.; Espinosa, A.; de Cid, R.; Garcia-Aymerich, J.; Carreras, A.; Cortés, B.; Pleguezuelos, V.; Jiménez, A.; et al. Ambient Air Pollution in Relation to SARS-CoV-2 Infection, Antibody Response, and COVID-19 Disease: A Cohort Study in Catalonia, Spain (COVICAT Study). Environ. Health Perspect. 2021, 129, 117003. [Google Scholar] [CrossRef]
- Bozack, A.; Pierre, S.; DeFelice, N.; Colicino, E.; Jack, D.; Chillrud, S.N.; Rundle, A.; Astua, A.; Quinn, J.W.; McGuinn, L.; et al. Long-Term Air Pollution Exposure and COVID-19 Mortality: A Patient-Level Analysis from New York City. Am. J. Respir. Crit. Care Med. 2022, 205, 651–662. [Google Scholar] [CrossRef]
- Di Ciaula, A.; Bonfrate, L.; Portincasa, P.; Appice, C.; Belfiore, A.; Binetti, M.; Cafagna, G.; Campanale, G.; Carrieri, A.; Cascella, G.; et al. Nitrogen Dioxide Pollution Increases Vulnerability to COVID-19 through Altered Immune Function. Environ. Sci. Pollut. Res. 2022, 29, 44404–44412. [Google Scholar] [CrossRef]
- López-Feldman, A.; Heres, D.; Marquez-Padilla, F. Air Pollution Exposure and COVID-19: A Look at Mortality in Mexico City Using Individual-Level Data. Sci. Total Environ. 2021, 756, 143929. [Google Scholar] [CrossRef]
- Mendy, A.; Wu, X.; Keller, J.L.; Fassler, C.S.; Apewokin, S.; Mersha, T.B.; Xie, C.; Pinney, S.M. Long-Term Exposure to Fine Particulate Matter and Hospitalization in COVID-19 Patients. Respir. Med. 2021, 178, 106313. [Google Scholar] [CrossRef]
- Mendy, A.; Wu, X.; Keller, J.L.; Fassler, C.S.; Apewokin, S.; Mersha, T.B.; Xie, C.; Pinney, S.M. Air Pollution and the Pandemic: Long-Term PM2.5 Exposure and Disease Severity in COVID-19 Patients. Respirology 2021, 26, 1181–1187. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J.; Kwong, J.; Kim, J.; van Donkelaar, A.; Martin, R.V.; Hystad, P.; Su, Y.; Lavigne, E.; Kirby-McGregor, M.; et al. Association between Long-Term Exposure to Ambient Air Pollution and COVID-19 Severity: A Prospective Cohort Study. CMAJ 2022, 194, E693–E700. [Google Scholar] [CrossRef]
- Bergamaschi, R.; Ponzano, M.; Schiavetti, I.; Carmisciano, L.; Cordioli, C.; Filippi, M.; Radaelli, M.; Immovilli, P.; Capobianco, M.; De Rossi, N.; et al. The Effect of Air Pollution on COVID-19 Severity in a Sample of Patients with Multiple Sclerosis. Eur. J. Neurol. 2022, 29, 535–542. [Google Scholar] [CrossRef]
- Bowe, B.; Xie, Y.; Gibson, A.K.; Cai, M.; van Donkelaar, A.; Martin, R.V.; Burnett, R.; Al-Aly, Z. Ambient Fine Particulate Matter Air Pollution and the Risk of Hospitalization among COVID-19 Positive Individuals: Cohort Study. Environ. Int. 2021, 154, 106564. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.; Bodinier, B.; Whitaker, M.; Delpierre, C.; Vermeulen, R.; Tzoulaki, I.; Elliott, P.; Chadeau-Hyam, M. COVID-19 Mortality in the UK Biobank Cohort: Revisiting and Evaluating Risk Factors. Eur. J. Epidemiol. 2021, 36, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Konstantinoudis, G.; Padellini, T.; Bennett, J.; Davies, B.; Ezzati, M.; Blangiardo, M. Long-Term Exposure to Air-Pollution and COVID-19 Mortality in England: A Hierarchical Spatial Analysis. Environ. Int. 2021, 146, 106316. [Google Scholar] [CrossRef] [PubMed]
- Pegoraro, V.; Heiman, F.; Levante, A.; Urbinati, D.; Peduto, I. An Italian Individual-Level Data Study Investigating on the Association between Air Pollution Exposure and COVID-19 Severity in Primary-Care Setting. BMC Public Health 2021, 21, 902. [Google Scholar] [CrossRef]
- Marquès, M.; Correig, E.; Ibarretxe, D.; Anoro, E.; Antonio Arroyo, J.; Jericó, C.; Borrallo, R.M.; Miret, M.; Näf, S.; Pardo, A.; et al. Long-Term Exposure to PM10 above WHO Guidelines Exacerbates COVID-19 Severity and Mortality. Environ. Int. 2022, 158, 106930. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, B.Z.; Sidell, M.A.; Chow, T.; Eckel, S.P.; Pavlovic, N.; Martinez, M.P.; Lurmann, F.; Thomas, D.C.; Gilliland, F.D.; et al. Near-Roadway Air Pollution Associated with COVID-19 Severity and Mortality—Multiethnic Cohort Study in Southern California. Environ. Int. 2021, 157, 106862. [Google Scholar] [CrossRef]
- Gangwar, R.S.; Bevan, G.H.; Palanivel, R.; Das, L.; Rajagopalan, S. Oxidative Stress Pathways of Air Pollution Mediated Toxicity: Recent Insights. Redox Biol. 2020, 34, 101545. [Google Scholar] [CrossRef]
- Yan, Z.; Jin, Y.; An, Z.; Liu, Y.; Samet, J.M.; Wu, W. Inflammatory Cell Signaling Following Exposures to Particulate Matter and Ozone. Biochim. Biophys. Acta BBA—Gen. Subj. 2016, 1860, 2826–2834. [Google Scholar] [CrossRef]
- Manivannan, J.; Sundaresan, L. Systems Level Insights into the Impact of Airborne Exposure on SARS-CoV-2 Pathogenesis and COVID-19 Outcome—A Multi-Omics Big Data Study. Gene Rep. 2021, 25, 101312. [Google Scholar] [CrossRef]
- Woodby, B.; Arnold, M.M.; Valacchi, G. SARS-CoV-2 Infection, COVID-19 Pathogenesis, and Exposure to Air Pollution: What Is the Connection? Ann. N. Y. Acad. Sci. 2021, 1486, 15–38. [Google Scholar] [CrossRef]
- Signorini, A.; Segre, A.M.; Polgreen, P.M. The Use of Twitter to Track Levels of Disease Activity and Public Concern in the U.S. during the Influenza A H1N1 Pandemic. PLoS ONE 2011, 6, e19467. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-I.; Tsai, C.-H.; Sun, Y.-L.; Hsieh, W.-Y.; Lin, Y.-C.; Chen, C.-Y.; Lin, C.-S. Instillation of Particulate Matter 2.5 Induced Acute Lung Injury and Attenuated the Injury Recovery in ACE2 Knockout Mice. Int. J. Biol. Sci. 2018, 14, 253–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.-H.; Liu, C.-C.; Hsu, T.-W.; Lin, J.-H.; Hsu, J.-W.; Li, A.F.-Y.; Yeh, Y.-C.; Hung, S.-C.; Hsu, H.-S. Upregulation of ACE2 and TMPRSS2 by Particulate Matter and Idiopathic Pulmonary Fibrosis: A Potential Role in Severe COVID-19. Part. Fibre Toxicol. 2021, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Steptoe, A.; Feldman, P.J. Neighborhood Problems as Sources of Chronic Stress: Development of a Measure of Neighborhood Problems, and Associations with Socioeconomic Status and Health. Ann. Behav. Med. 2001, 23, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Zajacova, A.; Dowd, J.B.; Aiello, A.E. Socioeconomic and Race/Ethnic Patterns in Persistent Infection Burden among U.S. Adults. J. Gerontol. Ser. A 2009, 64, 272–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowd, J.B.; Zajacova, A.; Aiello, A. Early Origins of Health Disparities: Burden of Infection, Health, and Socioeconomic Status in U.S. Children. Soc. Sci. Med. 2009, 68, 699–707. [Google Scholar] [CrossRef] [Green Version]
- Yaya, S.; Bishwajit, G. Burden of Acute Respiratory Infections among Under-Five Children in Relation to Household Wealth and Socioeconomic Status in Bangladesh. Trop. Med. Infect. Dis. 2019, 4, 36. [Google Scholar] [CrossRef] [Green Version]
- Singleton, C.R.; Winkler, M.; Houghtaling, B.; Adeyemi, O.S.; Roehll, A.M.; Pionke, J.J.; Anderson Steeves, E. Understanding the Intersection of Race/Ethnicity, Socioeconomic Status, and Geographic Location: A Scoping Review of U.S. Consumer Food Purchasing. Int. J. Environ. Res. Public. Health 2020, 17, 7677. [Google Scholar] [CrossRef]
- Belanger, M.J.; Hill, M.A.; Angelidi, A.M.; Dalamaga, M.; Sowers, J.R.; Mantzoros, C.S. COVID-19 and Disparities in Nutrition and Obesity. N. Engl. J. Med. 2020, 383, e69. [Google Scholar] [CrossRef]
- Rehm, C.D.; Peñalvo, J.L.; Afshin, A.; Mozaffarian, D. Dietary Intake among US Adults, 1999-2012. JAMA 2016, 315, 2542–2553. [Google Scholar] [CrossRef] [Green Version]
- Wallace, J.M.; Vaughn, M.G.; Bachman, J.G.; O’Malley, P.M.; Johnston, L.D.; Schulenberg, J.E. Race/Ethnicity, Socioeconomic Factors, and Smoking among Early Adolescent Girls in the United States. Drug Alcohol Depend. 2009, 104, S42–S49. [Google Scholar] [CrossRef] [PubMed]
- Wagenknecht, L.E.; Perkins, L.L.; Cutter, G.R.; Sidney, S.; Burke, G.L.; Manolio, T.A.; Jacobs, D.R.; Liu, K.A.; Friedman, G.D.; Hughes, G.H. Cigarette Smoking Behavior Is Strongly Related to Educational Status: The CARDIA Study. Prev. Med. 1990, 19, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Dieteren, C.; Bonfrer, I. Socioeconomic Inequalities in Lifestyle Risk Factors across Low- and Middle-Income Countries. BMC Public Health 2021, 21, 951. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.A.; Taren, D. How Poverty Affects Diet to Shape the Microbiota and Chronic Disease. Nat. Rev. Immunol. 2018, 18, 279–287. [Google Scholar] [CrossRef]
- Miller, G.E.; Engen, P.A.; Gillevet, P.M.; Shaikh, M.; Sikaroodi, M.; Forsyth, C.B.; Mutlu, E.; Keshavarzian, A. Lower Neighborhood Socioeconomic Status Associated with Reduced Diversity of the Colonic Microbiota in Healthy Adults. PLoS ONE 2016, 11, e0148952. [Google Scholar] [CrossRef]
- Bowyer, R.C.E.; Jackson, M.A.; Le Roy, C.I.; Ni Lochlainn, M.; Spector, T.D.; Dowd, J.B.; Steves, C.J. Socioeconomic Status and the Gut Microbiome: A TwinsUK Cohort Study. Microorganisms 2019, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Foster, H.M.; Celis-Morales, C.A.; Nicholl, B.I.; Petermann-Rocha, F.; Pell, J.P.; Gill, J.M.; O’Donnell, C.A.; Mair, F.S. The Effect of Socioeconomic Deprivation on the Association between an Extended Measurement of Unhealthy Lifestyle Factors and Health Outcomes: A Prospective Analysis of the UK Biobank Cohort. Lancet Public Health 2018, 3, e576–e585. [Google Scholar] [CrossRef] [Green Version]
- Assari, S.; Chalian, H.; Bazargan, M. Race, Ethnicity, Socioeconomic Status, and Chronic Lung Disease in the U.S. Res. Health Sci. 2020, 5, 48–63. [Google Scholar] [CrossRef] [Green Version]
- Bello, A.K.; Peters, J.; Rigby, J.; Rahman, A.A.; Nahas, M.E. Socioeconomic Status and Chronic Kidney Disease at Presentation to a Renal Service in the United Kingdom. Clin. J. Am. Soc. Nephrol. 2008, 3, 1316–1323. [Google Scholar] [CrossRef] [Green Version]
- Guariguata, L.; Whiting, D.R.; Hambleton, I.; Beagley, J.; Linnenkamp, U.; Shaw, J.E. Global Estimates of Diabetes Prevalence for 2013 and Projections for 2035. Diabetes Res. Clin. Pract. 2014, 103, 137–149. [Google Scholar] [CrossRef]
- Gordon-Larsen, P.; Adair, L.S.; Popkin, B.M. The Relationship of Ethnicity, Socioeconomic Factors, and Overweight in U.S. Adolescents. Obes. Res. 2003, 11, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.; Jemal, A.; Cokkinides, V.; Singh, G.K.; Cardinez, C.; Ghafoor, A.; Thun, M. Cancer Disparities by Race/Ethnicity and Socioeconomic Status. CA. Cancer J. Clin. 2004, 54, 78–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, T.; Dassonville, C.; Derbez, M.; Ramalho, O.; Kirchner, S.; Crump, D.; Mandin, C. Relationships between Socioeconomic and Lifestyle Factors and Indoor Air Quality in French Dwellings. Environ. Res. 2015, 140, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Vanker, A.; Barnett, W.; Nduru, P.M.; Gie, R.P.; Sly, P.D.; Zar, H.J. Home Environment and Indoor Air Pollution Exposure in an African Birth Cohort Study. Sci. Total Environ. 2015, 536, 362–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, S.B.; Bruce, N.G.; Grigg, J.; Hibberd, P.L.; Kurmi, O.P.; Lam, K.H.; Mortimer, K.; Asante, K.P.; Balakrishnan, K.; Balmes, J.; et al. Respiratory Risks from Household Air Pollution in Low and Middle Income Countries. Lancet Respir. Med. 2014, 2, 823–860. [Google Scholar] [CrossRef] [Green Version]
- Singleton, R.; Salkoski, A.J.; Bulkow, L.; Fish, C.; Dobson, J.; Albertson, L.; Skarada, J.; Kovesi, T.; McDonald, C.; Hennessy, T.W.; et al. Housing Characteristics and Indoor Air Quality in Households of Alaska Native Children with Chronic Lung Conditions. Indoor Air 2017, 27, 478–486. [Google Scholar] [CrossRef]
- Ginsburg, A.S.; Izadnegahdar, R.; Berkley, J.A.; Walson, J.L.; Rollins, N.; Klugman, K.P. Undernutrition and Pneumonia Mortality. Lancet Glob. Health 2015, 3, e735–e736. [Google Scholar] [CrossRef] [Green Version]
- Holuka, C.; Merz, M.P.; Fernandes, S.B.; Charalambous, E.G.; Seal, S.V.; Grova, N.; Turner, J.D. The COVID-19 Pandemic: Does Our Early Life Environment, Life Trajectory and Socioeconomic Status Determine Disease Susceptibility and Severity? Int. J. Mol. Sci. 2020, 21, 5094. [Google Scholar] [CrossRef]
- Vliegenthart, J.; Noppe, G.; van Rossum, E.F.C.; Koper, J.W.; Raat, H.; van den Akker, E.L.T. Socioeconomic Status in Children Is Associated with Hair Cortisol Levels as a Biological Measure of Chronic Stress. Psychoneuroendocrinology 2016, 65, 9–14. [Google Scholar] [CrossRef]
- Cohen, S.; Doyle, W.J.; Baum, A. Socioeconomic Status Is Associated with Stress Hormones. Psychosom. Med. 2006, 68, 414–420. [Google Scholar] [CrossRef] [Green Version]
- Baum, A.; Garofalo, J.P.; Yali, A.M. Socioeconomic Status and Chronic Stress: Does Stress Account for SES Effects on Health? Ann. N. Y. Acad. Sci. 1999, 896, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Öhlin, B.; Nilsson, P.M.; Nilsson, J.-Å.; Berglund, G. Chronic Psychosocial Stress Predicts Long-Term Cardiovascular Morbidity and Mortality in Middle-Aged Men. Eur. Heart J. 2004, 25, 867–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopp, M.S.; Réthelyi, J. Where Psychology Meets Physiology: Chronic Stress and Premature Mortality—The Central-Eastern European Health Paradox. Brain Res. Bull. 2004, 62, 351–367. [Google Scholar] [CrossRef]
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mozaffarian, D.; Wilson, P.W.F.; Kannel, W.B. Beyond Established and Novel Risk Factors. Circulation 2008, 117, 3031–3038. [Google Scholar] [CrossRef] [Green Version]
- Barbaresko, J.; Rienks, J.; Nöthlings, U. Lifestyle Indices and Cardiovascular Disease Risk: A Meta-Analysis. Am. J. Prev. Med. 2018, 55, 555–564. [Google Scholar] [CrossRef]
- Shigeta, H.; Shigeta, M.; Nakazawa, A.; Nakamura, N.; Yoshikawa, T. Lifestyle, Obesity, and Insulin Resistance. Diabetes Care 2001, 24, 608. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.B. Globalization of Diabetes: The Role of Diet, Lifestyle, and Genes. Diabetes Care 2011, 34, 1249–1257. [Google Scholar] [CrossRef] [Green Version]
- Stengel, B.; Tarver-Carr, M.E.; Powe, N.R.; Eberhardt, M.S.; Brancati, F.L. Lifestyle Factors, Obesity and the Risk of Chronic Kidney Disease. Epidemiol. Camb. Mass 2003, 14, 479–487. [Google Scholar] [CrossRef]
- Hartman, J.E.; Boezen, H.M.; de Greef, M.H.; Bossenbroek, L.; ten Hacken, N.H. Consequences of Physical Inactivity in Chronic Obstructive Pulmonary Disease. Expert Rev. Respir. Med. 2010, 4, 735–745. [Google Scholar] [CrossRef]
- Gobbens, R.J.J.; van Assen, M.A.L.M.; Luijkx, K.G.; Wijnen-Sponselee, M.T.; Schols, J.M.G.A. Determinants of Frailty. J. Am. Med. Dir. Assoc. 2010, 11, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, S.; Voortman, T.; Kiefte-de Jong, J.C.; van Rooij, F.J.A.; Ikram, M.A.; Rivadeneira, F.; Franco, O.H.; Schoufour, J.D. The Association between Lifestyle and Overall Health, Using the Frailty Index. Arch. Gerontol. Geriatr. 2018, 76, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babszky, G.; Torma, F.; Aczel, D.; Bakonyi, P.; Gombos, Z.; Feher, J.; Szabó, D.; Ligeti, B.; Pongor, S.; Balogh, L.; et al. COVID-19 Infection Alters the Microbiome: Elite Athletes and Sedentary Patients Have Similar Bacterial Flora. Genes 2021, 12, 1577. [Google Scholar] [CrossRef] [PubMed]
- Barraza-Villarreal, A.; Sunyer, J.; Hernandez-Cadena, L.; Escamilla-Nuñez, M.C.; Sienra-Monge, J.J.; Ramírez-Aguilar, M.; Cortez-Lugo, M.; Holguin, F.; Diaz-Sánchez, D.; Olin, A.C.; et al. Air Pollution, Airway Inflammation, and Lung Function in a Cohort Study of Mexico City Schoolchildren. Environ. Health Perspect. 2008, 116, 832–838. [Google Scholar] [CrossRef] [Green Version]
- Abramson, M.J.; Wigmann, C.; Altug, H.; Schikowski, T. Ambient Air Pollution Is Associated with Airway Inflammation in Older Women: A Nested Cross-Sectional Analysis. BMJ Open Respir. Res. 2020, 7, e000549. [Google Scholar] [CrossRef]
- Guarnieri, M.; Balmes, J.R. Outdoor Air Pollution and Asthma. Lancet 2014, 383, 1581–1592. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Sun, S.; Tang, R.; Qiu, H.; Huang, Q.; Mason, T.G.; Tian, L. Major Air Pollutants and Risk of COPD Exacerbations: A Systematic Review and Meta-Analysis. Int. J. Chron. Obstruct. Pulmon. Dis. 2016, 11, 3079–3091. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.S.V.; Langrish, J.P.; Nair, H.; McAllister, D.A.; Hunter, A.L.; Donaldson, K.; Newby, D.E.; Mills, N.L. Global Association of Air Pollution and Heart Failure: A Systematic Review and Meta-Analysis. Lancet Lond. Engl. 2013, 382, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Yorifuji, T.; Kashima, S.; Tsuda, T.; Ishikawa-Takata, K.; Ohta, T.; Tsuruta, K.; Doi, H. Long-Term Exposure to Traffic-Related Air Pollution and the Risk of Death from Hemorrhagic Stroke and Lung Cancer in Shizuoka, Japan. Sci. Total Environ. 2013, 443, 397–402. [Google Scholar] [CrossRef]
- Park, S.K.; Adar, S.D.; O’Neill, M.S.; Auchincloss, A.H.; Szpiro, A.; Bertoni, A.G.; Navas-Acien, A.; Kaufman, J.D.; Diez-Roux, A.V. Long-Term Exposure to Air Pollution and Type 2 Diabetes Mellitus in a Multiethnic Cohort. Am. J. Epidemiol. 2015, 181, 327–336. [Google Scholar] [CrossRef]
- Vogiatzis, I.; Zakynthinos, G.; Andrianopoulos, V. Mechanisms of Physical Activity Limitation in Chronic Lung Diseases. Pulm. Med. 2012, 2012, e634761. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, S.S.; Sima, C.A.; Kirkham, A.R.; Syed, N.; Camp, P.G. Physical Activity Measurement Accuracy in Individuals with Chronic Lung Disease: A Systematic Review with Meta-Analysis of Method Comparison Studies. Arch. Phys. Med. Rehabil. 2015, 96, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Gallant, J.; Hsue, P.Y.; Shreay, S.; Meyer, N. Comorbidities among US Patients with Prevalent HIV Infection—A Trend Analysis. J. Infect. Dis. 2017, 216, 1525–1533. [Google Scholar] [CrossRef]
- Perz, J.F.; Armstrong, G.L.; Farrington, L.A.; Hutin, Y.J.F.; Bell, B.P. The Contributions of Hepatitis B Virus and Hepatitis C Virus Infections to Cirrhosis and Primary Liver Cancer Worldwide. J. Hepatol. 2006, 45, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, T.P.; Rathnayaka, Y.; Perera, P.K.; Peachey, L.E.; Nolan, M.J.; Krause, L.; Rajakaruna, R.S.; Cantacessi, C. Infections by Human Gastrointestinal Helminths Are Associated with Changes in Faecal Microbiota Diversity and Composition. PLoS ONE 2017, 12, e0184719. [Google Scholar] [CrossRef]
- Stensvold, C.R.; van der Giezen, M. Associations between Gut Microbiota and Common Luminal Intestinal Parasites. Trends Parasitol. 2018, 34, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Giacomin, P.; Croese, J.; Krause, L.; Loukas, A.; Cantacessi, C. Suppression of Inflammation by Helminths: A Role for the Gut Microbiota? Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaiss, M.M.; Rapin, A.; Lebon, L.; Dubey, L.K.; Mosconi, I.; Sarter, K.; Piersigilli, A.; Menin, L.; Walker, A.W.; Rougemont, J.; et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity 2015, 43, 998–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human Gut Microbes Associated with Obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.W.; Bäckhed, F.; Landmesser, U.; Hazen, S.L. Intestinal Microbiota in Cardiovascular Health and Disease: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 2089–2105. [Google Scholar] [CrossRef] [PubMed]
- Trøseid, M.; Andersen, G.Ø.; Broch, K.; Hov, J.R. The Gut Microbiome in Coronary Artery Disease and Heart Failure: Current Knowledge and Future Directions. EBioMedicine 2020, 52, 102649. [Google Scholar] [CrossRef] [Green Version]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mähler, A.; Balogh, A.; Markó, L.; et al. Salt-Responsive Gut Commensal Modulates TH17 Axis and Disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Yang, T.; Santisteban, M.M.; Rodriguez, V.; Li, E.; Ahmari, N.; Carvajal, J.M.; Zadeh, M.; Gong, M.; Qi, Y.; Zubcevic, J.; et al. Gut Dysbiosis Is Linked to Hypertension. Hypertension 2015, 65, 1331–1340. [Google Scholar] [CrossRef] [Green Version]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The Influence of the Microbiome on Respiratory Health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.-Y.; Zhang, Z.; Feng, Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the Human Gut Microbiome in Liver Cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Mouries, J.; Brescia, P.; Silvestri, A.; Spadoni, I.; Sorribas, M.; Wiest, R.; Mileti, E.; Galbiati, M.; Invernizzi, P.; Adorini, L.; et al. Microbiota-Driven Gut Vascular Barrier Disruption Is a Prerequisite for Non-Alcoholic Steatohepatitis Development. J. Hepatol. 2019, 71, 1216–1228. [Google Scholar] [CrossRef]
- Ren, Z.; Fan, Y.; Li, A.; Shen, Q.; Wu, J.; Ren, L.; Lu, H.; Ding, S.; Ren, H.; Liu, C.; et al. Alterations of the Human Gut Microbiome in Chronic Kidney Disease. Adv. Sci. 2020, 7, 2001936. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Gut Instincts: Microbiota as a Key Regulator of Brain Development, Ageing and Neurodegeneration. J. Physiol. 2017, 595, 489–503. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.; Wu, X.; Hu, X.; Wang, T.; Jin, F. Recognizing Depression from the Microbiota–Gut–Brain Axis. Int. J. Mol. Sci. 2018, 19, 1592. [Google Scholar] [CrossRef] [Green Version]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The Healthy Human Microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, M.A.; Jeffery, I.B.; Beaumont, M.; Bell, J.T.; Clark, A.G.; Ley, R.E.; O’Toole, P.W.; Spector, T.D.; Steves, C.J. Signatures of Early Frailty in the Gut Microbiota. Genome Med. 2016, 8, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.E.; Kwak, S.E.; Lee, J.-H.; Zhang, D.; Bae, J.H.; Song, W. Exercise, the Gut Microbiome, and Frailty. Ann. Geriatr. Med. Res. 2019, 23, 105–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padilha de Lima, A.; Macedo Rogero, M.; Araujo Viel, T.; Garay-Malpartida, H.M.; Aprahamian, I.; Lima Ribeiro, S.M. Interplay between Inflammaging, Frailty and Nutrition in COVID-19: Preventive and Adjuvant Treatment Perspectives. J. Nutr. Health Aging 2022, 26, 67–76. [Google Scholar] [CrossRef]
- Bosco, N.; Noti, M. The Aging Gut Microbiome and Its Impact on Host Immunity. Genes Immun. 2021, 22, 289–303. [Google Scholar] [CrossRef]
- Ganesan, K.; Chung, S.K.; Vanamala, J.; Xu, B. Causal Relationship between Diet-Induced Gut Microbiota Changes and Diabetes: A Novel Strategy to Transplant Faecalibacterium Prausnitzii in Preventing Diabetes. Int. J. Mol. Sci. 2018, 19, 3720. [Google Scholar] [CrossRef] [Green Version]
- de Groot, P.F.; Belzer, C.; Aydin, Ö.; Levin, E.; Levels, J.H.; Aalvink, S.; Boot, F.; Holleman, F.; van Raalte, D.H.; Scheithauer, T.P.; et al. Distinct Fecal and Oral Microbiota Composition in Human Type 1 Diabetes, an Observational Study. PLoS ONE 2017, 12, e0188475. [Google Scholar] [CrossRef] [PubMed]
- Santisteban, M.M.; Qi, Y.; Zubcevic, J.; Kim, S.; Yang, T.; Shenoy, V.; Cole-Jeffrey, C.T.; Lobaton, G.O.; Stewart, D.C.; Rubiano, A.; et al. Hypertension-Linked Pathophysiological Alterations in the Gut. Circ. Res. 2017, 120, 312–323. [Google Scholar] [CrossRef]
- Jayashree, B.; Bibin, Y.S.; Prabhu, D.; Shanthirani, C.S.; Gokulakrishnan, K.; Lakshmi, B.S.; Mohan, V.; Balasubramanyam, M. Increased Circulatory Levels of Lipopolysaccharide (LPS) and Zonulin Signify Novel Biomarkers of Proinflammation in Patients with Type 2 Diabetes. Mol. Cell. Biochem. 2014, 388, 203–210. [Google Scholar] [CrossRef]
- Fulop, T.; Larbi, A.; Witkowski, J.M.; McElhaney, J.; Loeb, M.; Mitnitski, A.; Pawelec, G. Aging, Frailty and Age-Related Diseases. Biogerontology 2010, 11, 547–563. [Google Scholar] [CrossRef]
- Sewo Sampaio, P.Y.; Sampaio, R.A.C.; Coelho Júnior, H.J.; Teixeira, L.F.M.; Tessutti, V.D.; Uchida, M.C.; Arai, H. Differences in Lifestyle, Physical Performance and Quality of Life between Frail and Robust Brazilian Community-Dwelling Elderly Women. Geriatr. Gerontol. Int. 2016, 16, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.H.; Weiss, D.; Sourial, N.; Karunananthan, S.; Quail, J.M.; Wolfson, C.; Bergman, H. Frailty and Its Association with Disability and Comorbidity in a Community-Dwelling Sample of Seniors in Montreal: A Cross-Sectional Study. Aging Clin. Exp. Res. 2010, 22, 54–62. [Google Scholar] [CrossRef]
- Espinoza, S.E.; Quiben, M.; Hazuda, H.P. Distinguishing Comorbidity, Disability, and Frailty. Curr. Geriatr. Rep. 2018, 7, 201–209. [Google Scholar] [CrossRef]
- Foley, M.K.; Searle, S.D.; Toloue, A.; Booth, R.; Falkenham, A.; Falzarano, D.; Rubino, S.; Francis, M.E.; McNeil, M.; Richardson, C.; et al. Centenarians and Extremely Old People Living with Frailty Can Elicit Durable SARS-CoV-2 Spike Specific IgG Antibodies with Virus Neutralization Functions Following Virus Infection as Determined by Serological Study. eClinicalMedicine 2021, 37, 100975. [Google Scholar] [CrossRef]
- Seiffert, P.; Konka, A.; Kasperczyk, J.; Kawa, J.; Lejawa, M.; Maślanka-Seiffert, B.; Zembala-John, J.; Bugdol, M.; Romanik, M.; Bułdak, R.; et al. Immunogenicity of the BNT162b2 MRNA COVID-19 Vaccine in Older Residents of a Long-Term Care Facility: Relation with Age, Frailty and Prior Infection Status. Biogerontology 2022, 23, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Salmerón Ríos, S.; Mas Romero, M.; Cortés Zamora, E.B.; Tabernero Sahuquillo, M.T.; Romero Rizos, L.; Sánchez-Jurado, P.M.; Sánchez-Nievas, G.; Señalada, J.J.B.; García Nogueras, I.; Estrella Cazalla, J. de D.; et al. Immunogenicity of the BNT162b2 Vaccine in Frail or Disabled Nursing Home Residents: COVID-A Study. J. Am. Geriatr. Soc. 2021, 69, 1441–1447. [Google Scholar] [CrossRef]
- Dyer, A.H.; Noonan, C.; McElheron, M.; Batten, I.; Reddy, C.; Connolly, E.; Pierpoint, R.; Murray, C.; Leonard, A.; Higgins, C.; et al. Previous SARS-CoV-2 Infection, Age, and Frailty Are Associated with 6-Month Vaccine-Induced Anti-Spike Antibody Titer in Nursing Home Residents. J. Am. Med. Dir. Assoc. 2022, 23, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.R.; Sitaras, I.; Park, H.S.; Aytenfisu, T.Y.; Caputo, C.; Li, M.; Lee, J.; Johnston, T.S.; Li, H.; Wouters, C.; et al. Association of Frailty, Age, and Biological Sex with Severe Acute Respiratory Syndrome Coronavirus 2 Messenger RNA Vaccine-Induced Immunity in Older Adults. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2022, 75, S61–S71. [Google Scholar] [CrossRef] [PubMed]
- Frailty and Age Impact Immune Responses to Moderna COVID-19 MRNA Vaccine. Available online: https://www.researchsquare.com (accessed on 18 August 2022).
- Ferrucci, L.; Fabbri, E. Inflammageing: Chronic Inflammation in Ageing, Cardiovascular Disease, and Frailty. Nat. Rev. Cardiol. 2018, 15, 505–522. [Google Scholar] [CrossRef] [PubMed]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Alexander, H.D.; Ross, O.A. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef]
- Dowell, A.C.; Butler, M.S.; Jinks, E.; Tut, G.; Lancaster, T.; Sylla, P.; Begum, J.; Bruton, R.; Pearce, H.; Verma, K.; et al. Children Develop Robust and Sustained Cross-Reactive Spike-Specific Immune Responses to SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 40–49. [Google Scholar] [CrossRef]
- Cӑtoi, A.F.; Corina, A.; Katsiki, N.; Vodnar, D.C.; Andreicuț, A.D.; Stoian, A.P.; Rizzo, M.; Pérez-Martínez, P. Gut Microbiota and Aging-A Focus on Centenarians. Biochim. Biophys. Acta BBA—Mol. Basis Dis. 2020, 1866, 165765. [Google Scholar] [CrossRef]
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut Microbiome and Aging: Physiological and Mechanistic Insights. Nutr. Healthy Aging 2018, 4, 267–285. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Lertwattanarak, R.; Garduño, J.d.J.; Galeana, J.J.; Li, J.; Zamarripa, F.; Lancaster, J.L.; Mohan, S.; Hussey, S.; Musi, N. Elevated Muscle TLR4 Expression and Metabolic Endotoxemia in Human Aging. J. Gerontol. Ser. A 2015, 70, 232–246. [Google Scholar] [CrossRef]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A New Immune-Metabolic Viewpoint for Age-Related Diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef]
- Shintouo, C.M.; Mets, T.; Beckwee, D.; Bautmans, I.; Ghogomu, S.M.; Souopgui, J.; Leemans, L.; Meriki, H.D.; Njemini, R. Is Inflammageing Influenced by the Microbiota in the Aged Gut? A Systematic Review. Exp. Gerontol. 2020, 141, 111079. [Google Scholar] [CrossRef]
- Vaiserman, A.M.; Koliada, A.K.; Marotta, F. Gut Microbiota: A Player in Aging and a Target for Anti-Aging Intervention. Ageing Res. Rev. 2017, 35, 36–45. [Google Scholar] [CrossRef] [PubMed]
- MacSwain, K.L.H.; Sherry, S.B.; Stewart, S.H.; Watt, M.C.; Hadjistavropoulos, H.D.; Graham, A.R. Gender Differences in Health Anxiety: An Investigation of the Interpersonal Model of Health Anxiety. Personal. Individ. Differ. 2009, 47, 938–943. [Google Scholar] [CrossRef]
- Özdin, S.; Bayrak Özdin, Ş. Levels and Predictors of Anxiety, Depression and Health Anxiety during COVID-19 Pandemic in Turkish Society: The Importance of Gender. Int. J. Soc. Psychiatry 2020, 66, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Hiza, H.A.B.; Casavale, K.O.; Guenther, P.M.; Davis, C.A. Diet Quality of Americans Differs by Age, Sex, Race/Ethnicity, Income, and Education Level. J. Acad. Nutr. Diet. 2013, 113, 297–306. [Google Scholar] [CrossRef]
- Pivonello, R.; Auriemma, R.S.; Pivonello, C.; Isidori, A.M.; Corona, G.; Colao, A.; Millar, R.P. Sex Disparities in COVID-19 Severity and Outcome: Are Men Weaker or Women Stronger? Neuroendocrinology 2021, 111, 1066–1085. [Google Scholar] [CrossRef]
- Thompson, A.E.; Anisimowicz, Y.; Miedema, B.; Hogg, W.; Wodchis, W.P.; Aubrey-Bassler, K. The Influence of Gender and Other Patient Characteristics on Health Care-Seeking Behaviour: A QUALICOPC Study. BMC Fam. Pract. 2016, 17, 38. [Google Scholar] [CrossRef] [Green Version]
- Solomou, I.; Constantinidou, F. Prevalence and Predictors of Anxiety and Depression Symptoms during the COVID-19 Pandemic and Compliance with Precautionary Measures: Age and Sex Matter. Int. J. Environ. Res. Public. Health 2020, 17, 4924. [Google Scholar] [CrossRef]
- Tharakan, T.; Khoo, C.C.; Giwercman, A.; Jayasena, C.N.; Sofikitis, N.; Salonia, A.; Minhas, S. Are Sex Disparities in COVID-19 a Predictable Outcome of Failing Men’s Health Provision? Nat. Rev. Urol. 2022, 19, 47–63. [Google Scholar] [CrossRef]
- Cai, H. Sex Difference and Smoking Predisposition in Patients with COVID-19. Lancet Respir. Med. 2020, 8, e20. [Google Scholar] [CrossRef]
- Wilsnack, R.W.; Vogeltanz, N.D.; Wilsnack, S.C.; Harris, T.R. Gender Differences in Alcohol Consumption and Adverse Drinking Consequences: Cross-Cultural Patterns. Addiction 2000, 95, 251–265. [Google Scholar] [CrossRef]
- Aryal, S.; Diaz-Guzman, E.; Mannino, D.M. COPD and Gender Differences: An Update. Transl. Res. 2013, 162, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Kanter, R.; Caballero, B. Global Gender Disparities in Obesity: A Review. Adv. Nutr. 2012, 3, 491–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauvais-Jarvis, F. Gender Differences in Glucose Homeostasis and Diabetes. Physiol. Behav. 2018, 187, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-J.; Ma, Z.; Wang, J.; Chen, L.-X.; Zhong, J.-C. Gender Differences in Hypertension. J Cardiovasc. Transl. Res. 2020, 13, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Thom, T.; Haase, N.; Rosamond, W.; Howard, V.J.; Rumsfeld, J.; Manolio, T.; Zheng, Z.-J.; Flegal, K.; O’Donnell, C.; Kittner, S.; et al. Heart Disease and Stroke Statistics--2006 Update: A Report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2006, 113, e85–e151. [Google Scholar] [CrossRef] [Green Version]
- Wray, S.; Arrowsmith, S. The Physiological Mechanisms of the Sex-Based Difference in Outcomes of COVID19 Infection. Front. Physiol. 2021, 12, 627260. [Google Scholar] [CrossRef]
- Jazwinski, S.M. Longevity, Genes, and Aging. Science 1996, 273, 54–59. [Google Scholar] [CrossRef]
- Brooks-Wilson, A.R. Genetics of Healthy Aging and Longevity. Hum. Genet. 2013, 132, 1323–1338. [Google Scholar] [CrossRef] [Green Version]
- Forgetta, V.; Manousaki, D.; Istomine, R.; Ross, S.; Tessier, M.-C.; Marchand, L.; Li, M.; Qu, H.-Q.; Bradfield, J.P.; Grant, S.F.A.; et al. Rare Genetic Variants of Large Effect Influence Risk of Type 1 Diabetes. Diabetes 2020, 69, 784–795. [Google Scholar] [CrossRef] [Green Version]
- Hyttinen, V.; Kaprio, J.; Kinnunen, L.; Koskenvuo, M.; Tuomilehto, J. Genetic Liability of Type 1 Diabetes and the Onset Age among 22,650 Young Finnish Twin Pairs: A Nationwide Follow-Up Study. Diabetes 2003, 52, 1052–1055. [Google Scholar] [CrossRef] [Green Version]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research; Saxena, R.; Voight, B.F.; Lyssenko, V.; Burtt, N.P.; de Bakker, P.I.W.; Chen, H.; Roix, J.J.; Kathiresan, S.; Hirschhorn, J.N.; et al. Genome-Wide Association Analysis Identifies Loci for Type 2 Diabetes and Triglyceride Levels. Science 2007, 316, 1331–1336. [Google Scholar] [CrossRef]
- Scott, L.J.; Mohlke, K.L.; Bonnycastle, L.L.; Willer, C.J.; Li, Y.; Duren, W.L.; Erdos, M.R.; Stringham, H.M.; Chines, P.S.; Jackson, A.U.; et al. A Genome-Wide Association Study of Type 2 Diabetes in Finns Detects Multiple Susceptibility Variants. Science 2007, 316, 1341–1345. [Google Scholar] [CrossRef] [Green Version]
- Walley, A.J.; Asher, J.E.; Froguel, P. The Genetic Contribution to Non-Syndromic Human Obesity. Nat. Rev. Genet. 2009, 10, 431–442. [Google Scholar] [CrossRef]
- Yang, W.; Kelly, T.; He, J. Genetic Epidemiology of Obesity. Epidemiol. Rev. 2007, 29, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keating, M.T.; Sanguinetti, M.C. Molecular Genetic Insights into Cardiovascular Disease. Science 1996, 272, 681–685. [Google Scholar] [CrossRef]
- Genetic Variants in Novel Pathways Influence Blood Pressure and Cardiovascular Disease Risk. Nature 2011, 478, 103–109. [CrossRef] [PubMed] [Green Version]
- Kim, D.-K.; Weller, B.; Lin, C.-W.; Sheykhkarimli, D.; Knapp, J.J.; Dugied, G.; Zanzoni, A.; Pons, C.; Tofaute, M.J.; Maseko, S.B.; et al. A Proteome-Scale Map of the SARS-CoV-2–Human Contactome. Nat. Biotechnol. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.-H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable Escape of SARS-CoV-2 Omicron to Antibody Neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Satterfield, B.A.; Bhatt, D.L.; Gersh, B.J. Cardiac Involvement in the Long-Term Implications of COVID-19. Nat. Rev. Cardiol. 2022, 19, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xu, E.; Bowe, B.; Al-Aly, Z. Long-Term Cardiovascular Outcomes of COVID-19. Nat. Med. 2022, 28, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.; Meng, D.; Xiong, L.; Wu, S.; Yang, L.; Wang, S.; Zhou, M.; He, X.; Cao, X.; Xiong, H.; et al. Long-Term Effects of COVID-19 on Health Care Workers 1-Year Post-Discharge in Wuhan. Infect. Dis. Ther. 2022, 11, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.D.; Lavelle, M.; Boursiquot, B.C.; Wan, E.Y. Long-Term Complications of COVID-19. Am. J. Physiol.-Cell Physiol. 2022, 322, C1–C11. [Google Scholar] [CrossRef]
- Meyer, P.T.; Hellwig, S.; Blazhenets, G.; Hosp, J.A. Molecular Imaging Findings on Acute and Long-Term Effects of COVID-19 on the Brain: A Systematic Review. J. Nucl. Med. 2022, 63, 971–980. [Google Scholar] [CrossRef]
- Mehandru, S.; Merad, M. Pathological Sequelae of Long-Haul COVID. Nat. Immunol. 2022, 23, 194–202. [Google Scholar] [CrossRef]
- Taquet, M.; Sillett, R.; Zhu, L.; Mendel, J.; Camplisson, I.; Dercon, Q.; Harrison, P.J. Neurological and Psychiatric Risk Trajectories after SARS-CoV-2 Infection: An Analysis of 2-Year Retrospective Cohort Studies Including 1 284 437 Patients. Lancet Psychiatry 2022, 9, 815–827. [Google Scholar] [CrossRef]
- Unnikrishnan, R.; Misra, A. Diabetes and COVID19: A Bidirectional Relationship. Eur. J. Clin. Nutr. 2021, 75, 1332–1336. [Google Scholar] [CrossRef]
- Rey-Reñones, C.; Martinez-Torres, S.; Martín-Luján, F.M.; Pericas, C.; Redondo, A.; Vilaplana-Carnerero, C.; Dominguez, A.; Grau, M. Type 2 Diabetes Mellitus and COVID-19: A Narrative Review. Biomedicines 2022, 10, 2089. [Google Scholar] [CrossRef]
- Hsu, C. Yes, AKI Truly Leads to CKD. J. Am. Soc. Nephrol. 2012, 23, 967–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Aly, Z.; Xie, Y.; Bowe, B. High-Dimensional Characterization of Post-Acute Sequelae of COVID-19. Nature 2021, 594, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Raman, B.; Bluemke, D.A.; Lüscher, T.F.; Neubauer, S. Long COVID: Post-Acute Sequelae of COVID-19 with a Cardiovascular Focus. Eur. Heart J. 2022, 43, 1157–1172. [Google Scholar] [CrossRef]
- Carrillo-Garcia, P.; Garmendia-Prieto, B.; Cristofori, G.; Montoya, I.L.; Hidalgo, J.J.; Feijoo, M.Q.; Cortés, J.J.B.; Gómez-Pavón, J. Health Status in Survivors Older than 70 Years after Hospitalization with COVID-19: Observational Follow-up Study at 3 Months. Eur. Geriatr. Med. 2021, 12, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Greco, G.I.; Noale, M.; Trevisan, C.; Zatti, G.; Dalla Pozza, M.; Lazzarin, M.; Haxhiaj, L.; Ramon, R.; Imoscopi, A.; Bellon, S.; et al. Increase in Frailty in Nursing Home Survivors of Coronavirus Disease 2019: Comparison with Noninfected Residents. J. Am. Med. Dir. Assoc. 2021, 22, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, L.U.; Avelino-Silva, T.J.; Dias, M.B.; Jacob-Filho, W.; Aliberti, M.J.R.; on behalf of COVID-19 and Frailty (CO-FRAIL) Study Group and EPIdemiology of Critical COVID-19 (EPICCoV) Study Group, for COVID Hospital das Clinicas, University of Sao Paulo Medical School (HCFMUSP) Study Group. Patient-Centered Outcomes Following COVID-19: Frailty and Disability Transitions in Critical Care Survivors. Crit. Care Med. 2022, 50, 955–963. [Google Scholar] [CrossRef]
- Stockwell, S.; Trott, M.; Tully, M.; Shin, J.; Barnett, Y.; Butler, L.; McDermott, D.; Schuch, F.; Smith, L. Changes in Physical Activity and Sedentary Behaviours from before to during the COVID-19 Pandemic Lockdown: A Systematic Review. BMJ Open Sport Exerc. Med. 2021, 7, e000960. [Google Scholar] [CrossRef]
- Bu, F.; Bone, J.K.; Mitchell, J.J.; Steptoe, A.; Fancourt, D. Longitudinal Changes in Physical Activity during and after the First National Lockdown Due to the COVID-19 Pandemic in England. Sci. Rep. 2021, 11, 17723. [Google Scholar] [CrossRef]
- Ross, J.A.; Malone, P.K.; Levy, S. The Impact of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Pandemic on Substance Use in the United States. Clin. Infect. Dis. 2022, 75, S81–S85. [Google Scholar] [CrossRef]
- Calina, D.; Hartung, T.; Mardare, I.; Mitroi, M.; Poulas, K.; Tsatsakis, A.; Rogoveanu, I.; Docea, A.O. COVID-19 Pandemic and Alcohol Consumption: Impacts and Interconnections. Toxicol. Rep. 2021, 8, 529–535. [Google Scholar] [CrossRef]
- Sallie, S.N.; Ritou, V.; Bowden-Jones, H.; Voon, V. Assessing International Alcohol Consumption Patterns during Isolation from the COVID-19 Pandemic Using an Online Survey: Highlighting Negative Emotionality Mechanisms. BMJ Open 2020, 10, e044276. [Google Scholar] [CrossRef]
- McPhee, M.D.; Keough, M.T.; Rundle, S.; Heath, L.M.; Wardell, J.D.; Hendershot, C.S. Depression, Environmental Reward, Coping Motives and Alcohol Consumption During the COVID-19 Pandemic. Front. Psychiatry 2020, 11, 574676. [Google Scholar] [CrossRef] [PubMed]
- Josephson, A.; Kilic, T.; Michler, J.D. Socioeconomic Impacts of COVID-19 in Low-Income Countries. Nat. Hum. Behav. 2021, 5, 557–565. [Google Scholar] [CrossRef]
- Hamadani, J.D.; Hasan, M.I.; Baldi, A.J.; Hossain, S.J.; Shiraji, S.; Bhuiyan, M.S.A.; Mehrin, S.F.; Fisher, J.; Tofail, F.; Tipu, S.M.M.U.; et al. Immediate Impact of Stay-at-Home Orders to Control COVID-19 Transmission on Socioeconomic Conditions, Food Insecurity, Mental Health, and Intimate Partner Violence in Bangladeshi Women and Their Families: An Interrupted Time Series. Lancet Glob. Health 2020, 8, e1380–e1389. [Google Scholar] [CrossRef] [PubMed]
- Faghri, P.D.; Dobson, M.; Landsbergis, P.; Schnall, P.L. COVID-19 Pandemic: What Has Work Got to Do with It? J. Occup. Environ. Med. 2021, 63, e245. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Fan, W. Who Loses Income during the COVID-19 Outbreak? Evidence from China. Res. Soc. Stratif. Mobil. 2020, 68, 100522. [Google Scholar] [CrossRef]
- Picchioni, F.; Goulao, L.F.; Roberfroid, D. The Impact of COVID-19 on Diet Quality, Food Security and Nutrition in Low and Middle Income Countries: A Systematic Review of the Evidence. Clin. Nutr. 2021, 43, 2955–2964. [Google Scholar] [CrossRef]
- Krumer-Nevo, M.; Refaeli, T. COVID-19: A Poverty-Aware Perspective. Am. J. Orthopsychiatr. 2021, 91, 423–431. [Google Scholar] [CrossRef]
- Madai Boukar, A.; Mbock, O.; Kilolo, J.-M.M. The Impacts of the COVID-19 Pandemic on Employment in Cameroon: A General Equilibrium Analysis. Afr. Dev. Rev. 2021, 33, S88–S101. [Google Scholar] [CrossRef]
- Selden, T.M.; Berdahl, T.A. COVID-19 And Racial/Ethnic Disparities In Health Risk, Employment, And Household Composition: Study Examines Potential Explanations for Racial-Ethnic Disparities in COVID-19 Hospitalizations and Mortality. Health Aff. 2020, 39, 1624–1632. [Google Scholar] [CrossRef]
- Béné, C.; Bakker, D.; Chavarro, M.J.; Even, B.; Melo, J.; Sonneveld, A. Global Assessment of the Impacts of COVID-19 on Food Security. Glob. Food Secur. 2021, 31, 100575. [Google Scholar] [CrossRef]
- Agberotimi, S.F.; Akinsola, O.S.; Oguntayo, R.; Olaseni, A.O. Interactions Between Socioeconomic Status and Mental Health Outcomes in the Nigerian Context Amid COVID-19 Pandemic: A Comparative Study. Front. Psychol. 2020, 11, 559819. [Google Scholar] [CrossRef]
- Iob, E.; Frank, P.; Steptoe, A.; Fancourt, D. Levels of Severity of Depressive Symptoms among At-Risk Groups in the UK During the COVID-19 Pandemic. JAMA Netw. Open 2020, 3, e2026064. [Google Scholar] [CrossRef]
- Kikuchi, H.; Machida, M.; Nakamura, I.; Saito, R.; Odagiri, Y.; Kojima, T.; Watanabe, H.; Inoue, S. Development of Severe Psychological Distress among Low-Income Individuals during the COVID-19 Pandemic: Longitudinal Study. BJPsych Open 2021, 7, e50. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Thomsen, M.R.; Nayga, R.M. The Association between Food Insecurity and Mental Health during the COVID-19 Pandemic. BMC Public Health 2021, 21, 607. [Google Scholar] [CrossRef] [PubMed]
- Naylor-Wardle, J.; Rowland, B.; Kunadian, V. Socioeconomic Status and Cardiovascular Health in the COVID-19 Pandemic. Heart 2021, 107, 358–365. [Google Scholar] [CrossRef] [PubMed]
- George, A.; Stead, T.S.; Ganti, L. What’s the Risk: Differentiating Risk Ratios, Odds Ratios, and Hazard Ratios? Cureus 2020, 12, e10047. [Google Scholar] [CrossRef]

| Functional Category | Gene or Genetic Region | Severity | Function Related to COVID-19 Pathogenesis * | Sources # |
|---|---|---|---|---|
| Direct interaction with SARS-CoV-2 | ACE2 † | increased/decreased | facilitation of SARS-CoV-2 cell entry, regulation of cardiovascular and renal function | [63] |
| TMPRSS2 † | increased/decreased | facilitation of SARS-CoV-2 cell entry | [64,65,66] | |
| Respiratory surface barrier | MUC1 | increased | formation of respiratory mucosal barrier | [67,68] |
| MUC5B † | decreased | the major gel-forming mucin in mucus | [67,69] | |
| LZTFL1 †/(SLC6A20) | increased | regulation of protein trafficking to the ciliary membrane/(proline transportation in the kidney and small intestine) | [67,68,70,71,72] | |
| NAPSA/KCNC3 | increased | may be important in the processing of pulmonary surfactant protein B/mediates the voltage-dependent potassium ion permeability of excitable membranes | [68] | |
| Immunity | HLA region † | increased/decreased | recognition and presentation of tolerogen and immunogen protein epitopes | [64,67,68,71,73] |
| SFTPD | increased | innate immune protein in the lungs | [67] | |
| OAS1 †/(OAS3) | increased | innate cellular antiviral responses/(viral infection resistance) | [67,68,71,74,75] | |
| DPP9 | increased | role in MHC-I peptide presentation | [67,68,70,71] | |
| TYK2 | increased | Janus-kinase in cytokine signalization pathways | [67,68,71] | |
| IFNAR2 † | increased | interferon receptor formation for IFN-alpha and -beta | [67,68,70,71] | |
| TLR7 † | increased | recognition of single-stranded RNA viruses in the endosomal system | [67,76] | |
| DOCK2 † | increased | remodeling of the actin cytoskeleton required for migration in response to chemokine signaling in peripheral blood leukocytes | [77] | |
| Regulation of blood pressure | TAC4/KAT7 | increased | receptor activation → regulation of blood pressure, the immune system, and endocrine gland secretion/part of a complex with acetyltransferase activity | [68,71] |
| ACE1 † | increased/decreased | regulation of blood pressure and electrolyte levels | [64,65,78] | |
| Other | FOXP4 | increased | regulation of gene transcription on the cell and tissue levels | [67,68,71] |
| ELF5 † | increased | epithelium-specific gene regulation and differentiation of keratinocytes | [67,79] | |
| KANSL1/WNT3 | increased | role in histone acetylation → cell proliferation, mitosis/developmental regulation | [71] | |
| ABO † | increased/decreased | blood group determination | [68,80] | |
| ApoE † | increased | component of chylomicron, catabolism of triglycerides | [64,81] | |
| FBRLS1 | increased | neurological and non-neurological functions | [67] |
| Study Design | Outcome | N of Cases | Covariates | Effect | Source |
|---|---|---|---|---|---|
| Retrospective cohort | severe disease | 174,568 | age, race, ethnicity, insurance status, weight, BMI | aOR = 1.60 (1.51–1.69) | [120] |
| Retrospective cohort | mortality | 116,539 | age, presence of comorbidities | aOR = 1.42 (1.38–1.47) | [121] |
| Prospective cohort study | hospitalization | 16,475 | age, comorbidities, education level, income, work status | aHR = 1.63 (1.57–1.68) | [115] |
| Cross-sectional | ICU admission | 14,992 | age, race, ethnicity, marital status, insurance type, median income, BMI, smoking and 17 comorbidities | aOR = 1.39 (1.23–1.59) | [118] |
| Systematic review | critical outcome | 43,248 | - | pRR = 1.26 (1.17–1.36) | [122] |
| Systematic review | mortality | 423,117 | most studies collected information on covariates | pHR = 1.24 (1.07–1.41) | [123] |
| Systematic review | severe disease | 21,060 | 39 out of 41 studies adjusted for at least 3 covariates | pOR = 1.51 (1.33–1.71) | [124] |
| Systematic review | severe disease | ~440,000 | most studies collected information on covariates | pOR = 2.05 (1.39–3.04) | [11] |
| Category | Comorbidity | Covariates | Effect | Source |
|---|---|---|---|---|
| respiratory | chronic obstructive lung disease (COPD) | age, sex, income, urbanization, LTC residency, comorbidities | HR = 1.19 (1.12–1.26) | [201] |
| age, sex, socioeconomic status, ethnicity, health care service, comorbidities | OR = 1.35 (1.27–1.42) | [202] | ||
| Variable * | OR = 1.25 (1.08–1.34) | [203] | ||
| cardiovascular | hypertension | age, sex, income, urbanization, LTC residency, comorbidities | HR = 1.16 (1.07–1.26) | [201] |
| age, sex, socioeconomic status, ethnicity, health care service, comorbidities | OR = 1.39 (1.35–1.44) | [202] | ||
| variable * | RR = 1.42 (1.30–1.54) | [204] | ||
| cardiovascular disease (CVD) | age, sex, income, urbanization, LTC residency, comorbidities | HR = 1.22 (1.15–1.30) | [201] | |
| age, sex, socioeconomic status, ethnicity, health care service, comorbidities | OR = 1.06 (1.02–1.12) | [202] | ||
| variable * | OR = 3.11 (2.55–3.79) | [205] | ||
| renal | chronic kidney disease (CKD) | age, sex, income, urbanization, LTC residency, comorbidities | HR = 1.45 (1.34–1.57) | [201] |
| age, sex, socioeconomic status, ethnicity, health care service, comorbidities | OR = 2.40 (2.30–2.51) | [202] | ||
| variable * | OR = 5.81 (3.78–8.94) | [206] | ||
| metabolic | diabetes (DM) | age, sex, income, urbanization, long-term care (LTC) residency, comorbidities | HR = 1.19 (1.12–1.26) | [201] |
| age, sex, socioeconomic status, ethnicity, health care service, comorbidities | OR = 1.82 (1.76–1.88) | [202] | ||
| variable * | RR = 1.54 (1.44–1.64) | [204] | ||
| obesity | age, sex, socioeconomic status, ethnicity, health care service, comorbidities | OR = 1.68 (1.62–1.73) | [202] | |
| variable * | RR = 1.45 (1.31–1.61) | [204] | ||
| variable * | OR = 1.61 (1.29–2.01) | [207] | ||
| other | cancer | age, sex, income, urbanization, LTC residency, comorbidities | HR = 1.17 (1.09–1.27) | [201] |
| variable * | OR = 1.71 (1.539–1.905) | [208] | ||
| variable * | RR = 1.44 (1.19–1.76) | [209] |
| Comorbidity | Studies Reporting Correlation with COVID-19 Outcome | Studies Reporting Lack of Correlation with COVID-19 Outcome | |
|---|---|---|---|
| asthma | death [247] | Severity * [269], hospitalization * [239,270], ICU * [239], death * [202,208,239,271,272,273,274,275], severity [276], hospitalization [203,277,278], ICU [203,277,278], MV [274,278], death [13,201,203,270,277,278,279,280,281] | |
| interstitial lung disease (ILD) | severity [231], ICU [232], death [232,233,234] | hospitalization [232], MV [232,233] | |
| coronary heart disease (CHD) | severity [124,282], ICU [283], death [283,284] | death [285,286] | |
| chronic liver disease (CLD) | hospitalization [287,288], severity [120,124,289,290], ICU [291], MV [291], death [208,280,281,287,288,292] | severity [293,294], death [201,280,291,295] | |
| liver cirrhosis | severity [296], death [287,296,297,298,299] | severity [296,300], death [300] | |
| metabolic associated/non-alcoholic fatty liver disease (MAFLD/NAFLD) | severity [301,302,303,304,305,306,307,308], ICU [291,308], MV [291], death [309] | ICU [307], death [291,308,309,310] | |
| alcohol-related liver disease (ALD) | death [299,310] | severity [299] | |
| immune-mediated inflammatory disease (IMID)/autoimmune disease | hospitalization [311], severity [312] | severity [313,314], MV [315], death [313,314,315,316,317] | |
| rheumatoid arthritis (RA) | hospitalization [288], severity [201,290], death [280,281,288] | severity [318,319], ICU [319], MV [319], death [201,318,319] | |
| immunosuppression | hospitalization [320,321], severity [322], ICU [323], death [13,280,281,321,324,325,326] | hospitalization [320,321], severity [327], MV [315], death [315,321,323,326] | |
| organ transplant | hospitalization [328], severity [269,329], ICU [323,330], death [201,280,281,329,331] | severity [332,333], death [323,330,334,335,336,337] | |
| asplenia | death [281] | death [280,281,338] | |
| cognitive disorder | severity [339], death [339] | - | |
| dementia | hospitalization [340], severity [120,341], death [201,280,286,339,340,341,342,343,344,345,346,347,348] | severity [339], ICU [349] | |
| Alzheimer’s disease (AD) | severity [350,351], death [342,343,344,350,352] | MV [351] | |
| cerebrovascular disease (CeVD) | severity [124,269,290,293,353,354,355,356,357], ICU [355,358], MV [358], death [354,355,356,358] | - | |
| stroke | severity [359], death [280,359,360,361] | severity [339], death [339] | |
| epilepsy | severity [362], death [339] | severity [339], death [362] | |
| obstructive sleep apnea (OSA) | hospitalization [346], severity [290,363,364,365], ICU [363], MV [363], death [363,366] | MV [367], death [367] | |
| Parkinson’s disease (PD) | severity [350] | hospitalization [368], death [342,350,369] | |
| mood disorders | hospitalization [370], severity [339], death [339,370,371,372] | severity [370] | |
| bipolar disorder | hospitalization [288,373], severity [374], death [288,344,373,374,375] | - | |
| major depressive disorder/depression | hospitalization [288,370,376,377], severity [365,374], death [286,288,344,370,374,376] | hospitalization [378], severity [339,370], death [371] | |
| psychotic disorders | hospitalization [379,380], death [339,371,376,380] | hospitalization [376], severity [339], death [344] | |
| schizophrenia | severity [374], death [339,374,381,382,383] | hospitalization [378], severity [339] | |
| stress-related disorder | - | hospitalization [376], death [339,376] | |
| substance use disorders | hospitalization [376,379,384,385], MV [384], death [371,384] | ICU [385], death [344,376,385] | |
| attention deficit hyperactivity disorder (ADHD) | severity [339,378], death [378] | death [339] | |
| Group | Species | Potential Factors Facilitating Co/Superinfection | Effect | Potential Mechanisms * |
|---|---|---|---|---|
| Bacteria | Pneumonia causing bacteria # | dysbiosis [426], disrupted epithelial barrier [427], hyperactive immune response [427], NET degradation [428], mechanical ventilation [429] | increased severity [430,431,432,433]not associated [434,435,436] | ↑ exacerbation of inflammation [437,438] and pneumonia [439,440,441], ↑ reduced T cell, B cell and mucosal IgA responses [437] |
| Mycobacterium tuberculosis † | increased attachment and colonization due to weakened immunity [442] | increased severity [443,444,445]not associated [446,447] | ↑ exacerbation of inflammation [442,448], ↑ upregulated IFN responses [449,450], ↑ depletion of immune cells targeting MT [442,448,451], ↑ interference with SARS-CoV-2-specific immunity [452], ↓ heterologous immunity [446], ↓ lower risk of immune-mediated damage [448] | |
| Viruses | HBV # | increased HBV reactivation in immunosuppressed patients [453], but not in general [454] | increased severity [455]not associated [443,456] | ↑ higher risk of liver injury [455,457,458,459], ↓ suppression of overactive immune responses [460] |
| HCV † | both utilize structurally similar ion channels [461] | not associated [462] | ↑ heightened inflammation [463,464], ↑ vascular endothelial dysfunction [465], ↑ extrahepatic damage [466], ↑ liver cirrhosis [467] | |
| HIV † | uncontrolled infection [468] | increased severity ‡ [445,469,470,471,472,473,474,475]not associated [443] | ↑ uncontrolled infection [468,471,476,477,478] with reduced B cell functions [479,480,481,482,483], lymphopenia [451,471,476,478,484], chronic inflammation [485,486] and comorbidities [477,487] | |
| influenza viruses † | interferon-induced overexpression of ACE2 [488,489] | increased severity [490,491]not associated [492] | ↑ increased inflammation [493,494,495,496], ↓ viral interference through antibodies [492,497,498,499], or interferon effects [500,501] | |
| HRV † | a HRV serotype overexpresses ACE2 and TMPRSS2 on epithelial cells [502] | not associated [503,504] | ↓ induced epithelial IFN responses block SARS-CoV-2 replication [505,506] | |
| Fungi | Aspergillus spp. # | dysregulated immune system (corticosteroids, lymphopenia) [507,508] | high reported CFR [509,510] | ↑ exacerbation of pneumonia (IL-6 [511,512], IL-10 [513,514]) |
| Candida spp. # | dysregulated immune system (corticosteroids, lymphopenia) [507,508], mechanical ventilation [508], antibiotic use [508] | increased severity [431] | ↑ exacerbation of pneumonia [515] (IL-6 [516,517]) | |
| Parasites | Helminths † | altered mucus secretion [518] | reduced severity [519] | ↓ induced Th2 responses [520], ↓ attenuated sepsis [520], ↓ increased microbiota diversity [521,522], ↑ inability to produce early immune responses [520], ↑ nutritional and metabolic problems [523] |
| Entamoeba and Giardia spp. | - | reduced severity [519] | ↓ induced Th2 responses [522], ↓ increased diversity of microbiota or ↑ dysbiosis [522] | |
| Plasmodium spp. | - | increased severity [524] | ↑ T cell exhaustion [525] ↑ fewer atypical memory B cells [526], ↓ cross-reactivity [527] | |
| Trypanosoma spp. | - | not associated [528,529] | ↓chronic but regulated inflammation [528] |
| VOC | Reference | Covariates | Outcome | n | Effect (95% CI) | Source |
|---|---|---|---|---|---|---|
| Alpha | non-VOC | age, sex, ethnicity, index of multiple deprivation, lower tier local authority region, test date | death | 54,906 | aHR = 1.64 (1.32–2.04) | [861] |
| Alpha | non-VOC | age, sex, ethnicity, deprivation, residence in a care home, the local authority of residence, test date | death | 1,146,534 | aHR = 1.55 (1.39–1.72) | [862] |
| Probable Alpha (N501Y+) | non-VOC | age, sex, time, vaccination status, comorbidities, and pregnancy status | hospitalization | 162,854 | aOR = 1.52 (1.42–1.63) | [863] |
| ICU | aOR = 1.89 (1.67–2.17) | |||||
| death | aOR = 1.51 (1.30–1.78) | |||||
| Beta | non-VOC | age, sex, week of reporting, country | hospitalization | 436 | aOR = 3.6 (2.1–6.2) | [864] |
| ICU | aOR = 3.3 (1.9–5.7) | |||||
| death | aOR = 1.1 (0.4–3.4) | |||||
| Beta | Alpha | age, sex, diagnosis date | severe | 9182 | aOR = 1.24 (1.11–1.39) | [865] |
| critical | aOR = 1.49 (1.13–1.97) | |||||
| death | aOR = 1.57 (1.03–2.43) | |||||
| Gamma | non-VOC | age, sex, week of reporting, country | hospitalization | 352 | aOR = 4.2 (2.1–8.4) | [864] |
| ICU | aOR = 2.2 (1.8–2.9) | |||||
| death | aOR = 0.6 (0.3–1.0) | |||||
| Delta | non-VOC | age, sex, time, vaccination status, comorbidities, and pregnancy status | hospitalization | 5945 | aOR = 2.08 (1.78–2.40) | [863] |
| ICU | aOR = 3.35 (2.60–4.31) | |||||
| death | aOR = 2.33 (1.54–3.31) | |||||
| Delta | Alpha | age, sex, relative socioeconomic deprivation, ethnicity | hospitalization | 8682 | aHR = 2.26 (1.32–3.89) | [866] |
| hospitalization/emergency care | aHR = 1.45 (1.08–1.95) | |||||
| Delta | Alpha | age, sex, deprivation, test date, comorbidities | hospitalization | 9996 | aHR = 1.85 (1.39–2.47) | [867] |
| Omicron (BA.1) | Delta | sex, age, previous infection, vaccination status, Charlson comorbidity index | hospitalization | 6581 | aHR = 0.25 (0.15–0.43) | [868] |
| death | aHR = 0.14 (0.0011–1.12) | |||||
| Omicron (BA.1) | Delta | age, sex, race/ethnicity, and neighborhood-level median household income, smoking, body mass index, Charlson comorbidity index, health care utilization | hospitalization | 52,297 | aHR = 0.48 (0.36–0.64) | [869] |
| death | aHR = 0.09 (0.01–0.75) | |||||
| Omicron (BA.1) | Delta | age, sex, comorbidities, geography, vaccination, prior infection (corrected for under-ascertainment) | hospitalization/death | 5144 * | aHR = 0.72 | [870] |
| Omicron (BA.1) | Delta | reinfection (corrected for under-ascertainment of prior infections), vaccination status, 10-year age-band, sex, ethnicity, NHS region, specimen date | hospitalization | 55,583 | aHR = 0.65 | [675] |
| Omicron (BA.1) | Omicron (BA.2) | age, comorbidities, vaccination, ethnicity and race, sex, previous infection status | hospitalization | 1720 | aOR = 2.71 (2.42–3.02) | [871] |
| ICU | 232 | aOR = 3.06 (2.28–4.10) | ||||
| MV | 272 | aOR = 3.55 (2.61–4.84) | ||||
| death | 203 | aOR = 2.20 (1.56–3.11) | ||||
| Omicron (BA.2) | Omicron (BA.1) | age, sex, comorbidities, geography, health care sector, and previous SARS-CoV-2 infection | hospitalization | 8276 | aOR = 0·96 (0·85–1·09) | [872] |
| Omicron (BA.4/BA.5) | Omicron (BA.1) | age, sex, comorbidities, geography, health care sector | hospitalization | 1806 | aOR = 1.24 (0.98–1.55) | [674] |
| SES Indicator | Studies Reporting Correlation with COVID-19 Outcome | Studies Reporting Lack of Correlation with COVID-19 Outcome |
|---|---|---|
| Poverty | hospitalization [106,109,202], severe disease [916], length of stay [917], ICU [106], MV [917], mortality [109,202,280,281,912,918,919,920,921] | hospitalization [914,915], severe disease [922], ICU [912], MV [202], mortality [106,201,915] |
| Nutrition | severe disease [819,825], mortality [918] | severe disease [819,916] |
| Health care | severe disease [913], ICU [912], mortality [109,918] | mortality [912] |
| Education | mortality [918] | - |
| Minority status | hospitalization [106,109,914,917,923,924], severe disease [106,912,913,925], ICU [912], mortality [109,923,925] | hospitalization [923], ICU [106], mortality [106,912,915,921,923], |
| Housing | mortality [918] | severe disease [916] |
| Pollutant | Duration | Studies Reporting Association with COVID-19 Outcome | Studies Reporting Lack of Correlation with COVID-19 Outcome |
|---|---|---|---|
| B(a)P | Short term | mortality [935] | MV [935] |
| BC | Long term | - | severe disease [936], ICU [937], MV [937], mortality [937] |
| PM2.5 | Short term | mortality [935] | MV [935], mortality [938,939] |
| Long term | hospitalization [940,941,942], severe disease [936,943], ICU [937], mortality [937,939] | hospitalization [940,942,944], ICU [942], MV [937], mortality [942,945,946] | |
| PM10 | Short term | pneumonia [947] | MV [935], mortality [935] |
| Long term | severe disease [948], mortality [948] | mortality [945] | |
| NO2 | Short term | mortality [938] | - |
| Long term | severe disease [936] | mortality [937,942,946], hospitalization [942], ICU [937,942], MV [937], severe disease [948] | |
| NOx | Long term | ICU [949], mortality [949] | hospitalization [949], ICU [949], mortality [945,949] |
| O3 | Long term | hospitalization [942], ICU [942], mortality [942] | severe disease [936] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zsichla, L.; Müller, V. Risk Factors of Severe COVID-19: A Review of Host, Viral and Environmental Factors. Viruses 2023, 15, 175. https://doi.org/10.3390/v15010175
Zsichla L, Müller V. Risk Factors of Severe COVID-19: A Review of Host, Viral and Environmental Factors. Viruses. 2023; 15(1):175. https://doi.org/10.3390/v15010175
Chicago/Turabian StyleZsichla, Levente, and Viktor Müller. 2023. "Risk Factors of Severe COVID-19: A Review of Host, Viral and Environmental Factors" Viruses 15, no. 1: 175. https://doi.org/10.3390/v15010175
APA StyleZsichla, L., & Müller, V. (2023). Risk Factors of Severe COVID-19: A Review of Host, Viral and Environmental Factors. Viruses, 15(1), 175. https://doi.org/10.3390/v15010175