Characterization of Two Aggressive PepMV Isolates Useful in Breeding Programs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Virus Materials
2.2. Preparation of Infectious cDNA Clones
2.3. Virion Purification
2.4. Comparison of In Silico Sequences
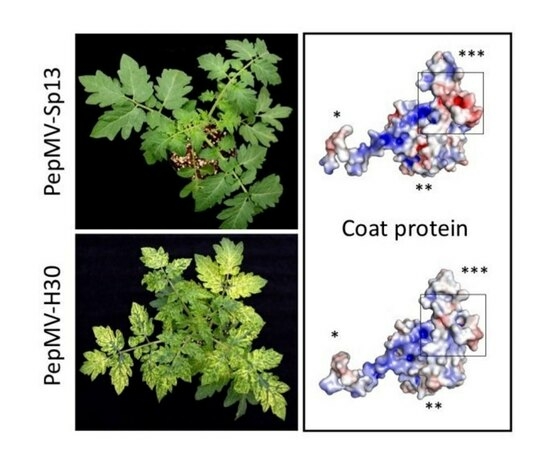
2.5. Structural Modeling of the PepMV-H30 CP
2.6. Site-Directed Mutagenesis
2.7. Confocal Laser Scanning Microscopy
3. Results
3.1. Characterization of Two PepMV Isolates Inducing Severe Bright Yellow Mosaics
3.2. Newly Described Mutations Associated with Yellow Mosaic Induction in N. benthamiana
3.3. Subcellular Localization of PepMV-EU Carrying Different Point Mutations in the CP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, R.A.C.; Koenig, R.; Lesemann, D.E. Pepino Mosaic Virus, a New Potexvirus from Pepino (Solanum muricatum). Ann. Appl. Biol. 1980, 94, 61–68. [Google Scholar] [CrossRef]
- Van der Vlugt, R.A.A.; Stijger, C.C.M.M.; Verhoeven, J.T.J.; Lesemann, D.E. First Report of Pepino Mosaic Virus on Tomato. Plant Dis. 2000, 84, 103. [Google Scholar] [CrossRef]
- Minicka, J.; Hasiów-Jaroszewska, B.; Borodynko-Filas, N.; Pospieszny, H.; Hanssen, I.M. Rapid Evolutionary Dynamics of the Pepino Mosaic Virus–Status and Future Perspectives. J. Plant Prot. Res. 2016, 56, 337–345. [Google Scholar] [CrossRef]
- Blystad, D.R.; van der Vlugt, R.; Alfaro-Fernández, A.; del Carmen Córdoba, M.; Bese, G.; Hristova, D.; Pospieszny, H.; Mehle, N.; Ravnikar, M.; Tomassoli, L.; et al. Host Range and Symptomatology of Pepino Mosaic Virus Strains Occurring in Europe. Eur. J. Plant Pathol. 2015, 143, 43–56. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Thomma, B.P. Pepino Mosaic Virus: A Successful Pathogen That Rapidly Evolved from Emerging to Endemic in Tomato Crops. Mol. Plant Pathol. 2010, 11, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, J.M.; Hernández-Gallardo, M.D.; Cenis, J.L.; Lacasa, A.; Aranda, M.A. Complete Sequence of the Pepino Mosaic Virus RNA Genome. Arch. Virol. 2002, 147, 2009–2015. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Solovyev, A.G. Triple Gene Block: Modular Design of a Multifunctional Machine for Plant Virus Movement. J. Gen. Virol. 2003, 84, 1351–1366. [Google Scholar] [CrossRef]
- Lough, T.J.; Emerson, S.J.; Lucas, W.J.; Forster, R.L.S. Trans-Complementation of Long-Distance Movement of White Clover Mosaic Virus Triple Gene Block (TGB) Mutants: Phloem-Associated Movement of TGBp1. Virology 2001, 288, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Mathioudakis, M.M.; Rodríguez-Moreno, L.; Navarro Sempere, R.; Aranda, M.A.; Livieratos, I. Multifaceted Capsid Proteins: Multiple Interactions Suggest Multiple Roles for Pepino Mosaic Virus Capsid Protein. Mol. Plant-Microbe Interact. 2014, 27, 1356–1369. [Google Scholar] [CrossRef]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and Trafficking of a Plant Virus Are Coupled at the Entrances of Plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Agirrezabala, X.; Méndez-López, E.; Lasso, G.; Sánchez-Pina, M.A.; Aranda, M.; Valle, M. The Near-Atomic CryoEM Structure of a Flexible Filamentous Plant Virus Shows Homology of Its Coat Protein with Nucleoproteins of Animal Viruses. eLife 2015, 4, e11795. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Cho, W.K.; Sohn, S.H.; Kim, K.H. Interaction of the Host Protein NbDnaJ with Potato Virus X Minus-Strand Stem-Loop 1 RNA and Capsid Protein Affects Viral Replication and Movement. Biochem. Biophys. Res. Commun. 2012, 417, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Park, M.R.; Kim, K.H. Molecular Characterization of the Interaction between the N-Terminal Region of Potato Virus X (PVX) Coat Protein (CP) and Nicotiana Benthamiana PVX CP-Interacting Protein, NbPCIP1. Virus Genes 2013, 46, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.S.; Nam, J.; Seo, E.Y.; Nam, M.; Vaira, A.M.; Bae, H.; Jang, C.Y.; Lee, C.H.; Kim, H.G.; Roh, M.; et al. The Coat Protein of Alternanthera Mosaic Virus Is the Elicitor of a Temperature-Sensitive Systemic Necrosis in Nicotiana Benthamiana, and Interacts with a Host Boron Transporter Protein. Virology 2014, 452–453, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Cho, W.K.; Kim, K.H. Two Homologous Host Proteins Interact with Potato Virus X RNAs and CPs and Affect Viral Replication and Movement. Sci. Rep. 2016, 6, 28743. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, A.G.; Makarov, V.V. Helical Capsids of Plant Viruses: Architecture with Structural Lability. J. Gen. Virol. 2016, 97, 1739–1754. [Google Scholar] [CrossRef]
- Lan, P.; Yen, W.B.; Tsai, C.W.; Lin, N.S. A Unique Glycine-Rich Motif at the N-Terminal Region of Bamboo Mosaic Virus Coat Protein Is Required for Symptom Expression. Mol. Plant-Microbe Interact. 2010, 23, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Méndez-López, E.; Donaire, L.; Gosálvez, B.; Díaz-Vivancos, P.; Sánchez-Pina, M.A.; Tilsner, J.; Aranda, M.A. Tomato SlGSTU38 Interacts with the PepMV Coat Protein and Promotes Viral Infection. New Phytol. 2023, 238, 332–348. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Veiga, R.; Ghita, M.; Tsikou, D.; Medina, V.; Canto, T.; Makris, A.M.; Livieratos, I.C. Pepino Mosaic Virus Capsid Protein Interacts with a Tomato Heat Shock Protein Cognate 70. Virus Res. 2012, 163, 28–39. [Google Scholar] [CrossRef]
- Candresse, T.; Marais, A.; Faure, C.; Dubrana, M.P.; Gombert, J.; Bendahmane, A. Multiple Coat Protein Mutations Abolish Recognition of Pepino Mosaic Potexvirus (PepMV) by the Potato Rx Resistance Gene in Transgenic Tomatoes. Mol. Plant-Microbe Interact. 2010, 23, 376–383. [Google Scholar] [CrossRef]
- Duff-Farrier, C.R.A.; Candresse, T.; Bailey, A.M.; Boonham, N.; Foster, G.D. Evidence for Different, Host-Dependent Functioning of Rx against Both Wild-Type and Recombinant Pepino Mosaic Virus. Mol. Plant Pathol. 2016, 17, 120–126. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Jackowiak, P.; Figlerowicz, M.; Pospieszny, H. Single Mutation Converts Mild Pathotype of the Pepino Mosaic Virus into Necrotic One. Virus Res. 2011, 159, 57–61. [Google Scholar] [CrossRef]
- Sempere, R.N.; Gomez-Aix, C.; Ruiz-Ramon, F.; Gomez, P.; Hasiów-Jaroszewska, B.; Sánchez Pina, M.A.; Aranda, M.A. Pepino Mosaic Virus RdRp-POL Domain Is a HR-like Elicitor Shared by Necrotic and Mild Isolates. Phytopathology 2016, 106, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Paeleman, A.; Ortega-Parra, N.; Borodynko, N.; Minicka, J.; Czerwoniec, A.; Thomma, B.P.H.J.; Hanssen, I.M. Ratio of Mutated versus Wild-Type Coat Protein Sequences in Pepino Mosaic Virus Determines the Nature and Severity of Yellowing Symptoms on Tomato Plants. Mol. Plant Pathol. 2013, 14, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Duff-Farrier, C.R.A.; Bailey, A.M.; Boonham, N.; Foster, G.D. A Pathogenicity Determinant Maps to the N-Terminal Coat Protein Region of the Pepino Mosaic Virus Genome. Mol. Plant Pathol. 2015, 16, 308–315. [Google Scholar] [CrossRef]
- Moreno-Perez, M.G.; Pagan, I.; Aragon-Caballero, L.; Caceres, F.; Fraile, A.; Garcia-Arenal, F. Ecological and Genetic Determinants of Pepino Mosaic Virus Emergence. J. Virol. 2014, 88, 3359–3368. [Google Scholar] [CrossRef] [PubMed]
- Gómez, P.; Sempere, R.; Aranda, M.A. Pepino Mosaic Virus and Tomato Torrado Virus: Two Emerging Viruses Affecting Tomato Crops in the Mediterranean Basin. In Viruses and Virus Diseases of Vegetables in the Mediterranean Basin; Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2012; Volume 84, pp. 505–532. [Google Scholar]
- Hanssen, I.M.; Paeleman, A.; Vandewoestijne, E.; Van Bergen, L.; Bragard, C.; Lievens, B.; Vanachter, A.C.R.C.; Thomma, B.P.H.J. Pepino Mosaic Virus Isolates and Differential Symptomatology in Tomato. Plant Pathol. 2009, 58, 450–460. [Google Scholar] [CrossRef]
- Minicka, J.; Elena, S.F.; Borodynko-Filas, N.; Rubiś, B.; Hasiów-Jaroszewska, B. Strain-Dependent Mutational Effects for Pepino Mosaic Virus in a Natural Host. BMC Evol. Biol. 2017, 17, 67. [Google Scholar] [CrossRef]
- Alcaide, C.; Rabadán, M.P.; Juárez, M.; Gómez, P. Long-Term Cocirculation of Two Strains of Pepino Mosaic Virus in Tomato Crops and Its Effect on Population Genetic Variability. Phytopathology 2020, 110, 49–57. [Google Scholar] [CrossRef]
- Blawid, R.; Nagata, T. Construction of an Infectious Clone of a Plant RNA Virus in a Binary Vector Using One-Step Gibson Assembly. J. Virol. Methods 2015, 222, 11–15. [Google Scholar] [CrossRef]
- Gibson, D.G. Synthesis of DNA Fragments in Yeast by One-Step Assembly of Overlapping Oligonucleotides. Nucleic Acids Res. 2009, 37, 6984–6990. [Google Scholar] [CrossRef]
- Gómez, P.; Sempere, R.N.; Elena, S.F.; Aranda, M.A. Mixed Infections of Pepino Mosaic Virus Strains Modulate the Evolutionary Dynamics of This Emergent Virus. J. Virol. 2009, 83, 12378–12387. [Google Scholar] [CrossRef]
- AbouHaidar, M.G.; Xu, H.; Hefferon, K.L. Potexvirus Isolation and RNA Extraction. In Plant Virology Protocols: From Virus Isolation to Transgenic Resistance; Humana Press: Totowa, NJ, USA, 1998; pp. 131–143. ISBN 978-1-59259-566-2. [Google Scholar]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER Server for Protein 3D Structure Prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Ferruz, N.; Schmidt, S.; Höcker, B. ProteinTools: A Toolkit to Analyze Protein Structures. Nucleic Acids Res. 2021, 49, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ramón, F.; Sempere, R.N.; Méndez-López, E.; Sánchez-Pina, M.A.; Aranda, M.A. Second Generation of Pepino Mosaic Virus Vectors: Improved Stability in Tomato and a Wide Range of Reporter Genes. Plant Methods 2019, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Agüero, J.; Gómez-Aix, C.; Sempere, R.N.; García-Villalba, J.; García-Núñez, J.; Hernando, Y.; Aranda, M.A. Stable and Broad Spectrum Cross-Protection against Pepino Mosaic Virus Attained by Mixed Infection. Front. Plant Sci. 2018, 9, 1810. [Google Scholar] [CrossRef] [PubMed]
- French, S.; Robson, B. What Is a Conservative Substitution? J. Mol. Evol. 1983, 19, 171–175. [Google Scholar] [CrossRef]
- Alcaide, C.; Aranda, M.A. Fluorescently Labelled Pepino Mosaic Virus Strains Share Infected Tissues and Colocalize in Cellular Aggregates Reminiscent of Viral Replication Complexes. Virus Res. 2023, 329, 199100. [Google Scholar] [CrossRef]
- Minicka, J.; Otulak, K.; Garbaczewska, G.; Pospieszny, H.; Hasiów-Jaroszewska, B. Ultrastructural Insights into Tomato Infections Caused by Three Different Pathotypes of Pepino Mosaic Virus and Immunolocalization of Viral Coat Proteins. Micron 2015, 79, 84–92. [Google Scholar] [CrossRef]
- Reinero, A.; Beachy, R.N. Reduced Photosystem II Activity and Accumulation of Viral Coat Protein in Chloroplasts of Leaves Infected with Tobacco Mosaic Virus. Plant Physiol. 1989, 89, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, Y.; Yang, M.; Liu, S.; Li, Z.; Wang, X.; Han, C.; Yu, J.; Li, D. The Barley Stripe Mosaic Virus Γb Protein Promotes Chloroplast-Targeted Replication by Enhancing Unwinding of RNA Duplexes. PLoS Pathog. 2017, 13, e1006319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, X.; Hong, Y.; Liu, Y. Chloroplast in Plant-Virus Interaction. Front. Microbiol. 2016, 7, 1565. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W.; Charlesworth, B.; Charlesworth, D.; Crow, J.F. Rates of Spontaneous Mutation. Genetics 1998, 148, 1667–1686. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.A.; Fraile, A.; Dopazo, J.; Malpica, J.M.; García-Arenal, F. Contribution of Mutation and RNA Recombination to the Evolution of a Plant Pathogenic RNA. J. Mol. Evol. 1997, 44, 81–88. [Google Scholar] [CrossRef]
- Sánchez-Campos, S.; Díaz, J.A.; Monci, F.; Bejarano, E.R.; Reina, J.; Navas-Castillo, J.; Aranda, M.A.; Moriones, E. High Genetic Stability of the Begomovirus Tomato Yellow Leaf Curl Sardinia Virus in Southern Spain over an 8-Year Period. Phytopathology 2002, 92, 842–849. [Google Scholar] [CrossRef]
- Sanjuán, R.; Domingo-Calap, P. Genetic Diversity and Evolution of Viral Populations. Encycl. Virol. 2021, 53–61. [Google Scholar] [CrossRef]
- Torre, C.; Donaire, L.; Gómez-Aix, C.; Juárez, M.; Peterschmitt, M.; Urbino, C.; Hernando, Y.; Agüero, J.; Aranda, M.A. Characterization of Begomoviruses Sampled during Severe Epidemics in Tomato Cultivars Carrying the Ty-1 Gene. Int. J. Mol. Sci. 2018, 19, 2614. [Google Scholar] [CrossRef] [PubMed]
- Pagán, I.; del Carmen Córdoba-Sellés, M.; Martínez-Priego, L.; Fraile, A.; Malpica, J.M.; Jordá, C.; García-Arenal, F. Genetic Structure of the Population of Pepino Mosaic Virus Infecting Tomato Crops in Spain. Phytopathology 2006, 96, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Aix, C.; Alcaide, C.; Agüero, J.; Faize, M.; Juárez, M.; Díaz-Marrero, C.J.; Botella-Guillén, M.; Espino, A.I.; Aranda, M.A.; Gómez, P. Genetic Diversity and Population Structure of Pepino Mosaic Virus in Tomato Crops of Spain and Morocco. Ann. Appl. Biol. 2019, 174, 284–292. [Google Scholar] [CrossRef]
- Pospieszny, H.; Borodynko, N. New Polish Isolate of Pepino Mosaic Virus Highly Distinct from European Tomato, Peruvian, and US2 Strains. Plant Dis. 2006, 90, 1106. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, I.M.; Paeleman, A.; Wittemans, L.; Goen, K.; Lievens, B.; Bragard, C.; Vanachter, A.C.R.C.; Thomma, B.P.H.J. Genetic Characterization of Pepino Mosaic Virus Isolates from Belgian Greenhouse Tomatoes Reveals Genetic Recombination. Eur. J. Plant Pathol. 2008, 121, 131–146. [Google Scholar] [CrossRef]
- Ling, K.S.; Wintermantel, W.M.; Bledsoe, M. Genetic Composition of Pepino Mosaic Virus Population in North American Greenhouse Tomatoes. Plant Dis. 2008, 92, 1683–1688. [Google Scholar] [CrossRef]
- Ruiz-Ramón, F.; Rodríguez-Sepúlveda, P.; Bretó, P.; Donaire, L.; Hernando, Y.; Aranda, M.A. The Tomato Calcium-Permeable Channel 4.1 (SlOSCA4.1) Is a Susceptibility Factor for Pepino Mosaic Virus. Plant Biotechnol. J. 2023, 1, 2140–2154. [Google Scholar] [CrossRef]
- Escriu, F.; Perry, K.L.; Garcia-Arenal, F. Transmissibility of Cucumber Mosaic Virus by Aphis Gossypii Correlates with Viral Accumulation and Is Affected by the Presence of Its Satellite RNA. Phytopathology 2000, 90, 1068–1072. [Google Scholar] [CrossRef]
- Wintermantel, W.M.; Cortez, A.A.; Anchieta, A.G.; Gulati-Sakhuja, A.; Hladky, L.L. Co-Infection by Two Criniviruses Alters Accumulation of Each Virus in a Host-Specific Manner and Influences Efficiency of Virus Transmission. Phytopathology 2008, 98, 1340–1345. [Google Scholar] [CrossRef]
- Froissart, R.; Doumayrou, J.; Vuillaume, F.; Alizon, S.; Michalakis, Y. The Virulence-Transmission Trade-off in Vector-Borne Plant Viruses: A Review of (Non-)Existing Studies. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, C.; Aranda, M.A. Determinants of Persistent Patterns of Pepino Mosaic Virus Mixed Infections. Front. Microbiol. 2021, 12, 694492. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; May, R.M. Coevolution of Hosts and Parasites. Parasitology 1982, 85, 411–426. [Google Scholar] [CrossRef]
- Ewald, P.W. Host-Parasite Relations, Vectors, and the Evolution of Disease Severity. Annu. Rev. Ecol. Syst. 1983, 14, 465–485. [Google Scholar] [CrossRef]
- Gómez, P.; Rodríguez-Hernández, A.M.; Moury, B.; Aranda, M. Genetic Resistance for the Sustainable Control of Plant Virus Diseases: Breeding, Mechanisms and Durability. Eur. J. Plant Pathol. 2009, 125, 1–22. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcaide, C.; Méndez-López, E.; Úbeda, J.R.; Gómez, P.; Aranda, M.A. Characterization of Two Aggressive PepMV Isolates Useful in Breeding Programs. Viruses 2023, 15, 2230. https://doi.org/10.3390/v15112230
Alcaide C, Méndez-López E, Úbeda JR, Gómez P, Aranda MA. Characterization of Two Aggressive PepMV Isolates Useful in Breeding Programs. Viruses. 2023; 15(11):2230. https://doi.org/10.3390/v15112230
Chicago/Turabian StyleAlcaide, Cristina, Eduardo Méndez-López, Jesús R. Úbeda, Pedro Gómez, and Miguel A. Aranda. 2023. "Characterization of Two Aggressive PepMV Isolates Useful in Breeding Programs" Viruses 15, no. 11: 2230. https://doi.org/10.3390/v15112230
APA StyleAlcaide, C., Méndez-López, E., Úbeda, J. R., Gómez, P., & Aranda, M. A. (2023). Characterization of Two Aggressive PepMV Isolates Useful in Breeding Programs. Viruses, 15(11), 2230. https://doi.org/10.3390/v15112230