Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa
Abstract
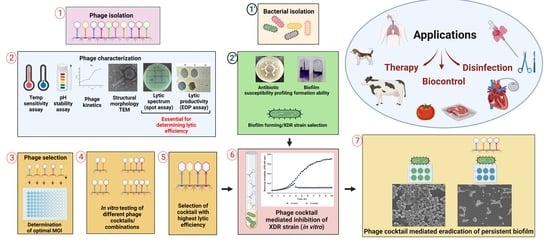
:1. Introduction
2. Materials and Methods
2.1. Bacterial Host Characterization
2.1.1. Antibiotic Resistance Profiling Using Disk Diffusion Method
2.1.2. Assessment of Biofilm Forming Ability
2.2. Isolation and Propagation of Bacteriophages
2.3. Characterization of Bacteriophages
2.3.1. Host Range and Cross Infectivity of Bacteriophages
2.3.2. Lytic Productivity of Bacteriophages by Efficiency of Plating (EOP)
- High productivity >0.5
- Medium productivity 0.5–0.1
- Low productivity 0.001–0.1
- Inefficient productivity <0.001
2.3.3. Bacteriophage Structural Morphology Determination by Transmission Electron Microscopy (TEM)
2.3.4. Phage Stability Assay over a Range of Temperature and pH
2.4. Microplate Single Phage or Cocktail Virulence Assay
2.4.1. Determination of Optimum Multiplicity of Infection (MOI) of Bacteriophages
2.4.2. Formulation and Assessment of Bacteriophage Cocktails in Liquid Infection Assay
2.5. Anti-Biofilm Assay of Bacteriophage Cocktail on Urinary Catheters and Borosilicate Glass
2.6. Statistical Analysis
3. Results
3.1. Host Bacterial Characterization and Selection of Biofilm-Forming XDR P. aeruginosa
3.2. Isolation and Characterization of Bacteriophages
3.3. Microplate Single Phage or Cocktail Virulence Assay
3.4. Anti-Biofilm Assay of Bacteriophage Cocktail on Urinary Catheters and Borosilicate Glass
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliver, A.; Mulet, X.; López-Causapé, C.; Juan, C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Update 2015, 21, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Al-Orphaly, M.; Hadi, H.A.; Eltayeb, F.K.; Al-Hail, H.; Samuel, B.G.; Sultan, A.A.; Skariah, S. Epidemiology of multidrug-resistant Pseudomonas aeruginosa in the Middle East and North Africa Region. mSphere 2021, 6, e00202-21. [Google Scholar] [CrossRef] [PubMed]
- Bodey, G.P.; Bolivar, R.; Fainstein, V.; Jadeja, L. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 1983, 5, 279–313. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, N.V.; Ausubel, F.M.; Ruvkun, G. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 1821–1826. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa Biofilms: Host Response and Clinical Implications in Lung Infections. Am. J. Respir. Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef]
- Yang, L.; Hu, Y.; Liu, Y.; Zhang, J.; Ulstrup, J.; Molin, S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 2011, 13, 1705–1717. [Google Scholar] [CrossRef]
- Musk, D.J., Jr.; Hergenrother, P.J. Chemical countermeasures for the control of bacterial biofilms: Effective compounds and promising targets. Curr. Med. Chem. 2006, 13, 2163–2177. [Google Scholar] [CrossRef] [Green Version]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Sønderholm, M.; Bjarnsholt, T.; Alhede, M.; Kolpen, M.; Jensen, P.Ø.; Kühl, M.; Kragh, K.N. The consequences of being in an infectious biofilm: Microenvironmental conditions governing antibiotic tolerance. Int. J. Mol. Sci. 2017, 18, 2688. [Google Scholar] [CrossRef]
- Wu, H.; Moser, C.; Wang, H.Z.; Høiby, N.; Song, Z.J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- Caldara, M.; Belgiovine, C.; Secchi, E.; Rusconi, R. Environmental, Microbiological, and Immunological Features of Bacterial Biofilms Associated with Implanted Medical Devices. Clin. Microbiol. Rev. 2022, 35, e0022120. [Google Scholar] [CrossRef]
- Sutherland, I.W.; Hughes, K.A.; Skillman, L.C.; Tait, K. The interaction of phage and biofilms. FEMS Microbiol. Lett. 2004, 232, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Glonti, T.; Chanishvili, N.; Taylor, P.W. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol. 2010, 108, 695–702. [Google Scholar] [CrossRef]
- Olszak, T.; Shneider, M.M.; Latka, A.; Maciejewska, B.; Browning, C.; Sycheva, L.V.; Cornelissen, A.; Danis-Wlodarczyk, K.; Senchenkova, S.N.; Shashkov, A.S.; et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Mi, L.; Liu, Y.; Wang, C.; He, T.; Gao, S.; Xing, S.; Huang, Y.; Fan, H.; Zhang, X.; Yu, W.; et al. Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Virus Genes 2019, 55, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liang, L.; Lin, S.; Jia, S. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 2010, 10, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielke, L.; Higgins, S.; Donoghue, A.; Donoghue, D.; Hargis, B.M. Salmonella host range of bacteriophages that infect multiple genera. Poult. Sci. 2007, 86, 2536–2540. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar]
- Pirnay, J.P.; De Vos, D.; Verbeken, G.; Merabishvili, M.; Chanishvili, N.; Vaneechoutte, M.; Zizi, M.; Laire, G.; Lavigne, R.; Huys, I.; et al. The phage therapy paradigm: Pret-a-porter or sur-mesure? Pharm. Res. 2011, 28, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Christiansen, R.H.; Dalsgaard, I.; Madsen, L.; Middelboe, M. Bacteriophage resistance mechanisms in the fish pathogen Flavobacterium psychrophilum: Linking genomic mutations to changes in bacterial virulence factors. Appl. Environ. Microbiol. 2015, 81, 1157–1167. [Google Scholar] [CrossRef]
- Alves, D.R.; Perez-Esteban, P.; Kot, W.; Bean, J.E.; Arnot, T.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb. Biotechnol. 2016, 9, 61–74. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; He, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 327. [Google Scholar] [CrossRef]
- Li, M.; Chang, R.Y.K.; Lin, Y.; Morales, S.; Kutter, E.; Chan, H.K. Phage cocktail powder for Pseudomonas aeruginosa respiratory infections. Int. J. Pharm. 2021, 596, 120200. [Google Scholar] [CrossRef]
- Hall, A.R.; De Vos, D.; Friman, V.P.; Pirnay, J.P.; Buckling, A. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl. Environ. Microbiol. 2012, 78, 5646–5652. [Google Scholar] [CrossRef]
- McVay, C.S.; Velásquez, M.; Fralick, J.A. Phage Therapy of Pseudomonas aeruginosa Infection in a Mouse Burn Wound Model. Antimicrob. Agents Chemother. 2007, 51, 1934–1938. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Änggård, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Mapes, A.C.; Trautner, B.W.; Liao, K.S.; Ramig, R.F. Development of expanded host range phage active on biofilms of multi-drug resistant Pseudomonas aeruginosa. Bacteriophage 2016, 6, e1096995. [Google Scholar] [CrossRef] [PubMed]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L.; et al. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef]
- Ong, S.P.; Azam, A.H.; Sasahara, T.; Miyanaga, K.; Tanji, Y. Characterization of Pseudomonas lytic phages and their application as a cocktail with antibiotics in controlling Pseudomonas aeruginosa. J. Biosci. Bioeng. 2020, 129, 693–699. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Bisno, A.L.; Beachey, E.H. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 1982, 37, 318–326. [Google Scholar] [CrossRef]
- Anand, T.; Bera, B.C.; Virmani, N.; Vaid, R.K.; Vashisth, M.; Tripathi, B.N. Isolation and characterization of a novel, T7-like phage against Aeromonas veronii. Virus Genes 2018, 54, 160–164. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Vettoretti, L.; Floret, N.; Hocquet, D.; Dehecq, B.; Plésiat, P.; Talon, D.; Bertrand, X. Emergence of extensive-drug-resistant Pseudomonas aeruginosa in a French university hospital. Eur. J. Clin. Microbiol. 2009, 28, 1217–1222. [Google Scholar] [CrossRef]
- Li, J.; Zou, M.; Dou, Q.; Hu, Y.; Wang, H.; Yan, Q.; Liu, W.E. Characterization of clinical extensively drug-resistant Pseudomonas aeruginosa in the Hunan province of China. Annal Clin. Microbiol. Antimicrob. 2016, 15, 1–7. [Google Scholar] [CrossRef]
- Aguilera-Sáez, J.; Andreu-Solà, V.; Escartín, N.L.; Garrido, V.R.; Gil, L.A.; García, J.S.; Campins, M.; Caparrós, J.B.; Barret, J.P. Extensively drug-resistant Pseudomonas aeruginosa outbreak in a burn unit: Management and solutions. Ann. Burns Fire Disasters 2019, 32, 47. [Google Scholar]
- Singh, S.; Pulusu, C.P.; Pathak, A.; Pradeep, B.E.; Prasad, K.N. Complete genome sequence of an extensively drug-resistant Pseudomonas aeruginosa ST773 clinical isolate from North India. J. Glob. Antimicrob. Resist. 2021, 27, 244–246. [Google Scholar] [CrossRef]
- Kracalik, I.; Ham, D.C.; McAllister, G.; Smith, A.R.; Vowles, M.; Kauber, K.; Zambrano, M.; Rodriguez, G.; Garner, K.; Chorbi, K.; et al. Extensively Drug-Resistant Carbapenemase-Producing Pseudomonas aeruginosa and Medical Tourism from the United States to Mexico, 2018–2019. Emerg. Infect. Dis. 2022, 28, 51. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–51. [Google Scholar] [CrossRef]
- Sahoo, J.P.; Mishra, A.P.; Samal, K.C.; Dash, A.K. Insights into the antibiotic resistance in Biofilms–A Review. Environ. Conserv. 2021, 22, 59–67. [Google Scholar] [CrossRef]
- Malhotra, R.; Dhawan, B.; Garg, B.; Shankar, V.; Nag, T.C. A Comparison of Bacterial Adhesion and Biofilm Formation on Commonly Used Orthopaedic Metal Implant Materials: An in vitro Study. Indian J. Orthop. 2019, 53, 148–153. [Google Scholar] [CrossRef]
- Alfa, M.J. Biofilms on instruments and environmental surfaces: Do they interfere with instrument reprocessing and surface disinfection? Review of the literature. Am. J. Infect. Control 2019, 47S, A39–A45. [Google Scholar] [CrossRef]
- Streicher, L.M. Exploring the future of infectious disease treatment in a post-antibiotic era: A comparative review of alternative therapeutics. J. Glob. Antimicrob. Resist. 2021, 24, 285–295. [Google Scholar] [CrossRef]
- Vieira, A.; Silva, Y.J.; Cunha, A.; Gomes, N.C.M.; Ackermann, H.W.; Almeida, A. Phage therapy to control multidrug-resistant Pseudomonas aeruginosa skin infections: In vitro and ex vivo experiments. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3241–3249. [Google Scholar] [CrossRef]
- Yuan, Y.; Qu, K.; Tan, D.; Li, X.; Wang, L.; Cong, C.; Xiu, Z.; Xu, Y. Isolation and characterization of a bacteriophage and its potential to disrupt multi-drug resistant Pseudomonas aeruginosa biofilms. Microb. Path. 2019, 128, 329–336. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Cafora, M.; Deflorian, G.; Forti, F.; Ferrari, L.; Binelli, G.; Briani, F.; Ghisotti, D.; Pistocchi, A. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci. Rep. 2019, 9, 1527. [Google Scholar] [CrossRef] [Green Version]
- Morello, E.; Saussereau, E.; Maura, D.; Huerre, M.; Touqui, L.; Debarbieux, L. Pulmonary bacteriophage therapy on Pseudomonas aeruginosa cystic fibrosis strains: First steps towards treatment and prevention. PLoS ONE 2011, 6, e16963. [Google Scholar] [CrossRef]
- Golkar, Z.; Bagasra, O.; Jamil, N. Experimental phage therapy on multiple drug resistant Pseudomonas aeruginosa infection in mice. J. Antivir. Antiretrovir. 2013, S10-005, 1–6. [Google Scholar] [CrossRef]
- Beeton, M.L.; Alves, D.R.; Enright, M.C.; Jenkins, A.T.A. Assessing phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int. J. Antimicrob Agents 2015, 46, 196–200. [Google Scholar] [CrossRef]
- Antoine, C.; Laforêt, F.; Blasdel, B.; Glonti, T.; Kutter, E.; Pirnay, J.P.; Mainil, J.; Delcenserie, V.; Thiry, D. Efficacy assessment of PEV2 phage on Galleria mellonella larvae infected with a Pseudomonas aeruginosa dog otitis isolate. Res. Vet. Sci. 2021, 136, 598–601. [Google Scholar] [CrossRef]
- Marza, J.A.S.; Soothill, J.S.; Boydell, P.; Collyns, T.A. Multiplication of therapeutically administered bacteriophages in Pseudomonas aeruginosa infected patients. Burns 2006, 32, 644–646. [Google Scholar] [CrossRef]
- Rhoads, D.D.; Wolcott, R.D.; Kuskowski, M.A.; Wolcott, B.M.; Ward, L.S.; Sulakvelidze, A. Bacteriophage therapy of venous leg ulcers in humans: Results of a phase I safety trial. J. Wound Care 2009, 18, 237–243. [Google Scholar] [CrossRef]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Valero-Rello, A. Diversity, specificity and molecular evolution of the lytic arsenal of Pseudomonas phages: In silico perspective. Environ. Microbiol. 2019, 21, 4136–4150. [Google Scholar] [CrossRef]
- Farlow, J.; Freyberger, H.R.; He, Y.; Ward, A.M.; Rutvisuttinunt, W.; Li, T.; Campbell, R.; Jacobs, A.C.; Nikolich, M.P.; Filippov, A.A. Complete genome sequences of 10 phages lytic against multidrug-resistant Pseudomonas aeruginosa. Microbiol. Resour. Announc. 2020, 9, e00503-20. [Google Scholar] [CrossRef]
- Kornienko, M.; Kuptsov, N.; Gorodnichev, R.; Bespiatykh, D.; Guliaev, A.; Letarova, M.; Kulikov, E.; Veselovsky, V.; Malakhova, M.; Letarov, A.; et al. Contribution of Podoviridae and Myoviridae bacteriophages to the effectiveness of anti-staphylococcal therapeutic cocktails. Sci. Rep. 2020, 10, 18612. [Google Scholar] [CrossRef] [PubMed]
- McCallin, S.; Sarker, S.A.; Barretto, C.; Sultana, S.; Berger, B.; Huq, S.; Krause, L.; Bibiloni, R.; Schmitt, B.; Reuteler, G.; et al. Safety analysis of a Russian phage cocktail: From metagenomic analysis to oral application in healthy human subjects. Virology 2013, 443, 187–196. [Google Scholar] [CrossRef]
- Camens, S.; Liu, S.; Hon, K.; Bouras, G.S.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Preclinical development of a bacteriophage cocktail for treating multidrug resistant Pseudomonas aeruginosa infections. Microorganisms 2021, 9, 2001. [Google Scholar] [CrossRef]
- Gill, J.J.; Hyman, P. Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.-P.; De Vos, D. Guidelines to Compose an Ideal Bacteriophage Cocktail. Methods Mol. Biol. 2018, 1693, 99–110. [Google Scholar] [PubMed]
- Abedon, S.T. Phage Therapy: Various Perspectives on How to Improve the Art. Methods Mol. Biol. 2018, 1734, 113–127. [Google Scholar]
- Yuan, Y.; Wang, L.; Li, X.; Tan, D.; Cong, C.; Xu, Y. Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii. Virus Res. 2019, 272, 197734. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Gelman, D.; Beyth, S.; Lerer, V.; Adler, K.; Poradosu-Cohen, R.; Coppenhagen-Glazer, S.; Hazan, R. Combined bacteriophages and antibiotics as an efficient therapy against VRE Enterococcus faecalis in a mouse model. Res. Microbiol. 2018, 169, 531–539. [Google Scholar] [CrossRef]
- Chen, L.; Yuan, S.; Liu, Q.; Mai, G.; Yang, J.; Deng, D.; Zhang, B.; Liu, C.; Ma, Y. In Vitro Design and Evaluation of Phage Cocktails Against Aeromonas salmonicida. Front. Microbiol. 2018, 9, 1476. [Google Scholar] [CrossRef]
- Gayder, S.; Parcey, M.; Nesbitt, D.; Castle, A.J.; Svircev, A.M. Population Dynamics between Erwinia amylovora, Pantoea agglomerans and Bacteriophages: Exploiting Synergy and Competition to Improve Phage Cocktail Efficacy. Microorganisms 2020, 8, 1449. [Google Scholar] [CrossRef]
- Kim, H.J.; Jun, J.W.; Giri, S.S.; Kim, S.G.; Kim, S.W.; Kwon, J.; Lee, S.B.; Chi, C.; Park, S.C. Bacteriophage cocktail for the prevention of multiple-antibiotic-resistant and mono-phage-resistant Vibrio coralliilyticus infection in pacific oyster (Crassostrea gigas) larvae. Pathogens 2020, 9, 831. [Google Scholar] [CrossRef]
- Donlan, R.M. Role of biofilms in antimicrobial resistance. ASAIO J. 2000, 46, S47–S52. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I.W. Polysaccharases for microbial exopolysaccharides. Carbohydr. Polym. 1991, 38, 319–328. [Google Scholar] [CrossRef]
- Yan, J.; Mao, J.; Xie, J. Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs 2014, 28, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Strength of Biofilm Formation | Number of Strains (Out of Total n = 29) | Percentage | Strains | Strength of Biofilm Formation as Visualized after CV Staining (Reference Images from Each Group) |
|---|---|---|---|---|
| Non-biofilm formers | 5 | 17.24% | VTCCBAA238, VTCCBAA239, VTCCBAA333, VTCCBAA843, VTCCBAA1096 |  |
| Weak biofilm formers | 2 | 06.89% | VTCCBAA951, VTCCBAA1216 |  |
| Moderate biofilm formers | 4 | 13.79% | VTCCBAA574, VTCCBAA632, VTCCBAA785, VTCCBAA1057 |  |
| Strong biofilm formers | 18 | 62.06% | VTCCBAA237, VTCCBAA325, VTCCBAA563, VTCCBAA789, VTCCBAA844, VTCCBAA845, VTCCBAA846, VTCCBAA848, VTCCBAA849, VTCCBAA956, VTCCBAA1061, VTCCBAA1047, VTCCBAA1097, RR/2021/112(571), Fop416A, Fop426A, Fop489B, Fop507C |  |
| Bacteriophages | φPA 170 | φPA 172 | φPA 173 | φPA 176 | φPA 177 | φPA 178 | φPA 180 |
|---|---|---|---|---|---|---|---|
| Total Pseudomonas strains used in the study (n = 29) | |||||||
| Strains lysed in spot test @ | 25 | 23 | 20 | 24 | 20 | 18 | 22 |
| Percent lysed strains β | 86% | 79% | 68% | 82% | 68% | 62% | 75% |
| High EOP # | 7 | 4 | 1 | 15 | 7 | 2 | 17 |
| Medium EOP $ | 4 | 1 | 1 | 3 | 0 | 1 | 4 |
| Low EOP * | 7 | 9 | 3 | 4 | 11 | 6 | 1 |
| Inefficient plating λ | 7 | 9 | 15 | 2 | 2 | 9 | 0 |
| Total EOP (=Total strains lysed) | |||||||
| High EOP/Total EOP | 0.28 | 0.17 | 0.05 | 0.63 | 0.35 | 0.11 | 0.77 |
| High+Medium EOP/Total EOP | 0.44 | 0.22 | 0.10 | 0.75 | 0.35 | 0.17 | 0.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vashisth, M.; Jaglan, A.B.; Yashveer, S.; Sharma, P.; Bardajatya, P.; Virmani, N.; Bera, B.C.; Vaid, R.K.; Anand, T. Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa. Viruses 2023, 15, 427. https://doi.org/10.3390/v15020427
Vashisth M, Jaglan AB, Yashveer S, Sharma P, Bardajatya P, Virmani N, Bera BC, Vaid RK, Anand T. Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa. Viruses. 2023; 15(2):427. https://doi.org/10.3390/v15020427
Chicago/Turabian StyleVashisth, Medhavi, Anu Bala Jaglan, Shikha Yashveer, Priya Sharma, Priyanka Bardajatya, Nitin Virmani, Bidhan Chand Bera, Rajesh Kumar Vaid, and Taruna Anand. 2023. "Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa" Viruses 15, no. 2: 427. https://doi.org/10.3390/v15020427
APA StyleVashisth, M., Jaglan, A. B., Yashveer, S., Sharma, P., Bardajatya, P., Virmani, N., Bera, B. C., Vaid, R. K., & Anand, T. (2023). Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa. Viruses, 15(2), 427. https://doi.org/10.3390/v15020427