Influence of Naturally Occurring Simian Foamy Viruses (SFVs) on SIV Disease Progression in the Rhesus Macaque (Macaca mulatta) Model
Abstract
:1. Introduction
2. Results and Discussion
2.1. Study Design

2.2. Evaluation of T and B cells


2.3. Plasma Viral Load Analysis in SIV-infected Monkeys

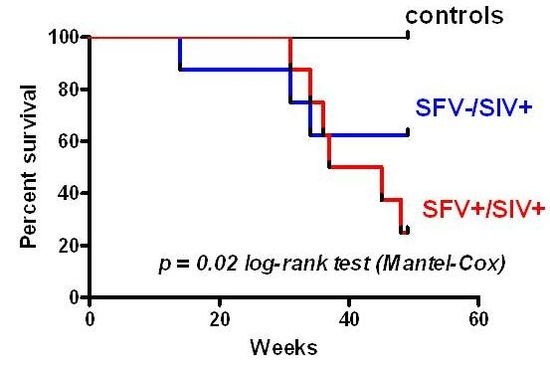
2.4. Survival Analysis and Disease Progression

| Monkey ID | SFV status | Disease progression (termination week) | Gp120 ELISA OD a (week 4 and week 37 or earlier terminal week) | ||
|---|---|---|---|---|---|
| Rapid (<6 month) | Conventional (6–24 month) | Slow (>2 year) | |||
| DBLM | − | 14 | 0.270/1.962 | ||
| DBFJ | + | 30 | NT b | ||
| DBNT | − | 31 | NT b | ||
| DBJD | − | 34 | NT b | ||
| DBN4 | + | 34 | 0.158/0.835 | ||
| DBHPA | + | 36 | NT b | ||
| DBHC | + | 36 | NT b | ||
| DBR7 | + | 45 | 0.235/1.294 | ||
| DBJM | + | 48 | 0.465/1.983 | ||
| DBJBA | + | 83 | 0.646/2.367 | ||
| DBJP | − | 83 | 0.356/1.475 | ||
| DBP8 | − | 115 | 0.167/1.049 | ||
| DBGK | − | 124 | 0.222/1.136 | ||
| DBC2 | − | 133 | 0.181/0.706 | ||
| DBGVc | − | 141 | 0.159/2.082 | ||
| DBP2 | + | 128 | 0.658/0.864 | ||
2.5. MHC Alleles

| Monkey ID | SFV status/Monkey survival (weeks) | Class I b | Class II b | ||
|---|---|---|---|---|---|
| Mamu-A | Mamu-B | Mamu-DR | Mamu-DP | ||
| A*01c A*02 A*08 | B*01 B*08 B*17 | B1*0306, B1*1003 B*w201, B*w606 B1*04 | B1*06 | ||
| DBLM | −/14 | A*02 | B*w201, B*w606 | ||
| DBFJ | +/30 | A*02 | B1*0306, B1*1003 B*w606 | ||
| DBNT | −/31 | A*01 A*08 | B*01 | B1*1003 B*w201, B*w606 | |
| DBJD | −/34 | A*02 A*08 | B*w201 B1*04 | ||
| DBN4 | +/34 | B*01 | B1*1003 B1*04 | B1*06 | |
| DBHPA | +/36 | A*02 | B*01 | B1*1003 | |
| DBHC | +/36 | B*01 | B1*1003 B*w201 B1*04 | ||
| DBR7 | +/45 | B1*0306 B1*1003 | |||
| DBJM | +/48 | A*02 | B*w606 B1*04 | ||
| DBJBA | +/83 | A*08 | B1*1003 | ||
| DBJP | −/83 | B1*1003 | |||
| DBP8 | −/115 | A*02 | B*w201, B*w606 | ||
| DBGK | −/124 | A*01 | B*01 | B1*0306, B1*1003 B*w201 | |
| DBC2 | −/133 | A*02 | B1*1003 B*w201, B*w606 | ||
| DBGV | −/141 | A*02 A*08 | B*w201, B*w606 | ||
| DBP2 | +/128 | B*08 B*17 | B1*1003 B*w201 | ||
3. Experimental
3.1. Animals, Prescreening for Selection, and Management
3.2. Preparation and Characterization of SIV Inoculum
3.3. Animal Injections, Blood Collections, and Necropsies
3.4. Flow Cytometry
3.5. Plasma Viral Load
3.6. Antibody Detection
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Johnson, P.R.; Hirsch, V.M. SIV infection of macaques as a model for AIDS pathogenesis. Int. Rev. Immunol. 1992, 8, 55–63. [Google Scholar]
- Murray, S.M.; Picker, L.J.; Axthelm, M.K.; Hudkins, K.; Alpers, C.E.; Linial, M.L. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J. Virol. 2008, 82, 5981–5985. [Google Scholar]
- Falcone, V.; Leupold, J.; Clotten, J.; Urbanyi, E.; Herchenroder, O.; Spatz, W.; Volk, B.; Bohm, N.; Toniolo, A.; Neumann-Haefelin, D.; et al. Sites of simian foamy virus persistence in naturally infected African green monkeys: Latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 1999, 257, 7–14. [Google Scholar]
- Switzer, W.M.; Salemi, M.; Shanmugam, V.; Gao, F.; Cong, M.E.; Kuiken, C.; Bhullar, V.; Beer, B.E.; Vallet, D.; Gautier-Hion, A.; et al. Ancient co-speciation of simian foamy viruses and primates. Nature 2005, 434, 376–380. [Google Scholar] [CrossRef]
- Liu, W.; Worobey, M.; Li, Y.; Keele, B.F.; Bibollet-Ruche, F.; Guo, Y.; Goepfert, P.A.; Santiago, M.L.; Ndjango, J.B.; Neel, C.; et al. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 2008, 4, e1000097. [Google Scholar] [CrossRef]
- Yu, S.F.; Stone, J.; Linial, M.L. Productive persistent infection of hematopoietic cells by human foamy virus. J. Virol. 1996, 70, 1250–1254. [Google Scholar]
- Saib, A.; Neves, M.; Giron, M.L.; Guillemin, M.C.; Valla, J.; Peries, J.; Canivet, M. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology 1997, 228, 263–268. [Google Scholar]
- Swack, N.S.; Hsiung, G.D. Pathogenesis of simian foamy virus infection in natural and experimental hosts. Infect Immun. 1975, 12, 470–474. [Google Scholar]
- Switzer, W.M.; Bhullar, V.; Shanmugam, V.; Cong, M.E.; Parekh, B.; Lerche, N.W.; Yee, J.L.; Ely, J.J.; Boneva, R.; Chapman, L.E.; et al. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates . J. Virol. 2004, 78, 2780–2789. [Google Scholar]
- Betsem, E.; Rua, R.; Tortevoye, P.; Froment, A.; Gessain, A. Frequent and recent human acquisition of simian foamy viruses through apes’ bites in central Africa. PLoS Pathog. 2011, 7, e1002306. [Google Scholar]
- Jones-Engel, L.; Engel, G.A.; Schillaci, M.A.; Rompis, A.; Putra, A.; Suaryana, K.G.; Fuentes, A.; Beer, B.; Hicks, S.; White, R.; et al. Primate-to-human retroviral transmission in Asia. Emerg. Infect. Dis. 2005, 11, 1028–1035. [Google Scholar]
- Wolfe, N.D.; Switzer, W.M.; Carr, J.K.; Bhullar, V.B.; Shanmugam, V.; Tamoufe, U.; Prosser, A.T.; Torimiro, J.N.; Wright, A.; Mpoudi-Ngole, E.; et al. Naturally acquired simian retrovirus infections in central African hunters. Lancet 2004, 363, 932–937. [Google Scholar]
- Khan, A.S. Simian foamy virus infection in humans: Prevalence and management. Expert Rev. Anti. Infect. Ther. 2009, 7, 569–580. [Google Scholar]
- Gessain, A.; Rua, R.; Betsem, E.; Turpin, J.; Mahieux, R. HTLV-3/4 and simian foamy retroviruses in humans: Discovery, epidemiology, cross-species transmission and molecular virology. Virology 2013, 435, 187–199. [Google Scholar]
- Boneva, R.S.; Switzer, W.M.; Spira, T.J.; Bhullar, V.B.; Shanmugam, V.; Cong, M.E.; Lam, L.; Heneine, W.; Folks, T.M.; Chapman, L.E. Clinical and virological characterization of persistent human infection with simian foamy viruses. AIDS Res. Hum. Retrovir. 2007, 23, 1330–1337. [Google Scholar]
- Switzer, W.M.; Garcia, A.D.; Yang, C.; Wright, A.; Kalish, M.L.; Folks, T.M.; Heneine, W. Coinfection with HIV-1 and simian foamy virus in West Central Africans. J. Infect. Dis. 2008, 197, 1389–1393. [Google Scholar]
- Murray, S.M.; Picker, L.J.; Axthelm, M.K.; Linial, M.L. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J. Virol. 2006, 80, 663–670. [Google Scholar]
- Keller, A.; Garrett, E.D.; Cullen, B.R. The Bel-1 protein of human foamy virus activates human immunodeficiency virus type 1 gene expression via a novel DNA target site. J. Virol. 1992, 66, 3946–3949. [Google Scholar]
- Marino, S.; Kretschmer, C.; Brandner, S.; Cavard, C.; Zider, A.; Briand, P.; Isenmann, S.; Wagner, E.F.; Aguzzi, A. Activation of HIV transcription by human foamy virus in transgenic mice. Lab. Invest. 1995, 73, 103–110. [Google Scholar]
- Schiffer, C.; Lecellier, C.H.; Mannioui, A.; Felix, N.; Nelson, E.; Lehmann-Che, J.; Giron, M.L.; Gluckman, J.C.; Saib, A.; Canque, B. Persistent infection with primate foamy virus type 1 increases human immunodeficiency virus type 1 cell binding via a Bet-independent mechanism. J. Virol. 2004, 78, 11405–11410. [Google Scholar]
- Giraldo-Vela, J.P.; Rudersdorf, R.; Chung, C.; Qi, Y.; Wallace, L.T.; Bimber, B.; Borchardt, G.J.; Fisk, D.L.; Glidden, C.E.; Loffredo, J.T.; et al. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J. Virol. 2008, 82, 859–870. [Google Scholar]
- Kaizu, M.; Borchardt, G.J.; Glidden, C.E.; Fisk, D.L.; Loffredo, J.T.; Watkins, D.I.; Rehrauer, W.M. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics 2007, 59, 693–703. [Google Scholar]
- Regier, D.A.; Desrosiers, R.C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 1990, 6, 1221–1231. [Google Scholar]
- Dang, Q.; Hirsch, V.M. Rapid disease progression to AIDS due to Simian immunodeficiency virus infection of macaques: Host and viral factors. Adv. Pharmacol. 2008, 56, 369–398. [Google Scholar]
- Montefiori, D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 2009, 485, 395–405. [Google Scholar]
- Bontrop, R.E.; Watkins, D.I. MHC polymorphism: AIDS susceptibility in non-human primates. Trends Immunol. 2005, 26, 227–233. [Google Scholar]
- Muhl, T.; Krawczak, M.; Ten Haaft, P.; Hunsmann, G.; Sauermann, U. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 2002, 169, 3438–3446. [Google Scholar]
- Mothe, B.R.; Weinfurter, J.; Wang, C.; Rehrauer, W.; Wilson, N.; Allen, T.M.; Allison, D.B.; Watkins, D.I. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 2003, 77, 2736–2740. [Google Scholar]
- Loffredo, J.T.; Maxwell, J.; Qi, Y.; Glidden, C.E.; Borchardt, G.J.; Soma, T.; Bean, A.T.; Beal, D.R.; Wilson, N.A.; Rehrauer, W.M.; et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 2007, 81, 8827–8832. [Google Scholar]
- Yant, L.J.; Friedrich, T.C.; Johnson, R.C.; May, G.E.; Maness, N.J.; Enz, A.M.; Lifson, J.D.; O'Connor, D.H.; Carrington, M.; Watkins, D.I. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 2006, 80, 5074–5077. [Google Scholar]
- Reed, J.S.; Sidney, J.; Piaskowski, S.M.; Glidden, C.E.; Leon, E.J.; Burwitz, B.J.; Kolar, H.L.; Eernisse, C.M.; Furlott, J.R.; Maness, N.J.; et al. The role of MHC class I allele Mamu-A*07 during SIV(mac)239 infection. Immunogenetics 2011, 63, 789–807. [Google Scholar]
- Heberling, R.L.; Kalter, S.S. Rapid dot-immunobinding assay on nitrocellulose for viral antibodies. J. Clin. Microbiol. 1986, 23, 109–113. [Google Scholar]
- Committee for the Update of the Guide for the Care and Use of Laboratory Animals; National Research Council. Guide for the Care and Use of Laboratory Animals; National Academy Press: Washington, DC, USA, 1996. [Google Scholar]
- Khan, A.S.; Sears, J.F.; Muller, J.; Galvin, T.A.; Shahabuddin, M. Sensitive assays for isolation and detection of simian foamy retroviruses. J. Clin. Microbiol. 1999, 37, 2678–2686. [Google Scholar]
- Newberg, M.H.; McEvers, K.J.; Gorgone, D.A.; Lifton, M.A.; Baumeister, S.H.; Veazey, R.S.; Schmitz, J.E.; Letvin, N.L. Immunodomination in the evolution of dominant epitope-specific CD8+ T lymphocyte responses in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 2006, 176, 319–328. [Google Scholar]
- Yuste, E.; Reeves, J.D.; Doms, R.W.; Desrosiers, R.C. Modulation of Env content in virions of simian immunodeficiency virus: Correlation with cell surface expression and virion infectivity. J. Virol. 2004, 78, 6775–6785. [Google Scholar]
- Karber, G.A. A contribution to the collective treatment of a pharmacological experimental series. Arch. Exp. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar]
- Salter, R.D.; Howell, D.N.; Cresswell, P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 1985, 21, 235–246. [Google Scholar]
- Sodora, D.L.; Lee, F.; Dailey, P.J.; Marx, P.A. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res. Hum. Retrovir. 1998, 14, 171–181. [Google Scholar]
- Dailey, P.J.; Zamroud, M.; Kelso, R.; Kolberg, J.; Urdea, M. Quantitation of Simian Immunodeficiency Virus (SIV) RNA in Plasma of Acute and Chronically Infected Rhesus Macaques Using a Branched DNA (bDNA) Signal Amplification Assay. In Proceedings of the 13th Annual Symposium Nonhuman Primate Models of AIDS, Monterey, CA, USA, 5–8 November 1995.
- Yuste, E.; Sanford, H.B.; Carmody, J.; Bixby, J.; Little, S.; Zwick, M.B.; Greenough, T.; Burton, D.R.; Richman, D.D.; Desrosiers, R.C.; et al. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: Replication, neutralization, and survey of HIV-1-positive plasma. J. Virol. 2006, 80, 3030–3041. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Choudhary, A.; Galvin, T.A.; Williams, D.K.; Beren, J.; Bryant, M.A.; Khan, A.S. Influence of Naturally Occurring Simian Foamy Viruses (SFVs) on SIV Disease Progression in the Rhesus Macaque (Macaca mulatta) Model. Viruses 2013, 5, 1414-1430. https://doi.org/10.3390/v5061414
Choudhary A, Galvin TA, Williams DK, Beren J, Bryant MA, Khan AS. Influence of Naturally Occurring Simian Foamy Viruses (SFVs) on SIV Disease Progression in the Rhesus Macaque (Macaca mulatta) Model. Viruses. 2013; 5(6):1414-1430. https://doi.org/10.3390/v5061414
Chicago/Turabian StyleChoudhary, Anil, Teresa A. Galvin, Dhanya K. Williams, Joel Beren, Mark A. Bryant, and Arifa S. Khan. 2013. "Influence of Naturally Occurring Simian Foamy Viruses (SFVs) on SIV Disease Progression in the Rhesus Macaque (Macaca mulatta) Model" Viruses 5, no. 6: 1414-1430. https://doi.org/10.3390/v5061414
APA StyleChoudhary, A., Galvin, T. A., Williams, D. K., Beren, J., Bryant, M. A., & Khan, A. S. (2013). Influence of Naturally Occurring Simian Foamy Viruses (SFVs) on SIV Disease Progression in the Rhesus Macaque (Macaca mulatta) Model. Viruses, 5(6), 1414-1430. https://doi.org/10.3390/v5061414