Nomenclature- and Database-Compatible Names for the Two Ebola Virus Variants that Emerged in Guinea and the Democratic Republic of the Congo in 2014
Abstract
:1. Introduction
2. Ebola Virus Strain, Variant, and Isolate Naming
<virus name>(/<strain>)/<isolation host-suffix>/<country of sampling>/<year of sampling>/<genetic variant designation>-<isolate designation>. [Note: the “/” between the <virus name> and the <isolation host-suffix> field was missing in the full name template outlined in [10], but is necessary for computational purposes. It is therefore introduced here and already implemented filovirus full names [11] will be retrospectively corrected.]
| Current Taxonomy and Nomenclature (Ninth ICTV Report and Updates) |
|---|
| Order Mononegavirales |
| Family Filoviridae |
| Genus Marburgvirus |
| Species Marburg marburgvirus |
| Virus 1: Marburg virus (MARV) |
| Virus 2: Ravn virus (RAVV) |
| Genus Ebolavirus |
| Species Taï Forest ebolavirus |
| Virus: Taï Forest virus (TAFV) |
| Species Reston ebolavirus |
| Virus: Reston virus (RESTV) |
| Species Sudan ebolavirus |
| Virus: Sudan virus (SUDV) |
| Species Zaire ebolavirus |
| Virus: Ebola virus (EBOV) |
| Species Bundibugyo ebolavirus |
| Virus: Bundibugyo virus (BDBV) |
| Genus Cuevavirus |
| Species Lloviu cuevavirus |
| Virus: Lloviu virus (LLOV) |
| full: | Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Ecran |
| shortened: | EBOV/H.sap/COD/76/Yam-Ecr |
| abbreviated: | EBOV/Yam-Ecr |
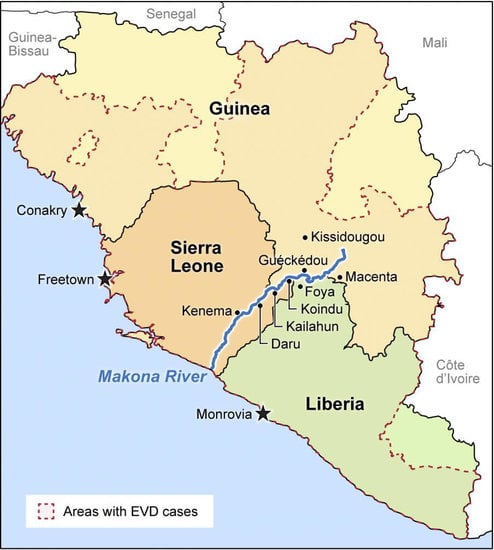
3. The 2014 Ebola Virus Variant Originating in Guinea


| Official short country name in English (geographical name) | Official country name in English (protocol name) | ISO 3166-1 3-letter abbreviation |
|---|---|---|
| France | French Republic | FRA |
| Germany | Federal Republic of Germany | DEU |
| Guinea | Republic of Guinea | GIN |
| LiberiaMaliNigeria | Republic of LiberiaRepublic of MaliFederal Republic of Nigeria | LBRMLINGA |
| NorwaySenegal | Kingdom of NorwayRepublic of Senegal | NORSEN |
| Sierra Leone | Republic of Sierra Leone | SLE |
| Spain | Kingdom of Spain | ESP |
| United Kingdom | United Kingdom of Great Britain and Northern Ireland | GBR |
| United States | United States of America | USA |
| GenBank Accession Number | <GenBank field> = Updated/corrected information |
|---|---|
| Final names | |
| KJ660346 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C15, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/GIN/2014/Makona-C15 | |
 | |
| KJ660347 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C07, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/GIN/2014/Makona-C07 | |
 | |
| KJ660348 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05 | |
 | |
| KM034549 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095B, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095B | |
 | |
| KM034550 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095 | |
 | |
| KM034551 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM096, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM096 | |
 | |
| KM034552 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM098, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM098 | |
 | |
| KM034553 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3670.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3670.1 | |
 | |
| KM034554 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.1 | |
 | |
| KM034555 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.2 | |
 | |
| KM034556 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.1 | |
 | |
| KM034557 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.2 | |
 | |
| KM034558 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3679.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3679.1 | |
 | |
| KM034559 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3680.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3680.1 | |
 | |
| KM034560 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3682.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3682.1 | |
 | |
| KM034561 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Mak-G3683.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Mak-G3683.1 | |
 | |
| KM034562 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Mak-G3686.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3686.1 | |
 | |
| KM034563 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3687.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3687.1 | |
 | |
| KM233035 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM104, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM104 | |
 | |
| KM233036 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM106, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM106 | |
 | |
| KM233037 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM110, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM110 | |
 | |
| KM233038 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM111, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM111 | |
 | |
| KM233039 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM112, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM112 | |
 | |
| KM233040 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM113, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM113 | |
 | |
| KM233041 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM115, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM115 | |
 | |
| KM233042 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM119, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM119 | |
 | |
| KM233043 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM120, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM120 | |
 | |
| KM233044 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM121, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM121 | |
 | |
| KM233045 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.1 | |
 | |
| KM233046 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.2 | |
 | |
| KM233047 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.3 | |
 | |
| KM233048 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.4, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.4 | |
 | |
| KM233049 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3707, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3707 | |
 | |
| KM233050 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.2 | |
 | |
| KM233051 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.3 | |
 | |
| KM233052 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.4, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.4 | |
 | |
| KM233053 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3724, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3724 | |
 | |
| KM233054 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3729, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3729 | |
 | |
| KM233055 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3734.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3734.1 | |
 | |
| KM233056 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.1 | |
 | |
| KM233057 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.2 | |
 | |
| KM233058 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.1 | |
 | |
| KM233059 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.2 | |
 | |
| KM233060 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.3 | |
 | |
| KM233061 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3752, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3752 | |
 | |
| KM233062 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3758, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3758 | |
 | |
| KM233063 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3764, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3764 | |
 | |
| KM233064 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3765.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3765.2 | |
 | |
| KM233065 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.1 | |
 | |
| KM233066 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.2 | |
 | |
| KM233067 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.3 | |
 | |
| KM233068 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.4, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.4 | |
 | |
| KM233069 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.1 | |
 | |
| KM233070 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.2 | |
 | |
| KM233071 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3771, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3771 | |
 | |
| KM233072 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3782, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3782 | |
 | |
| KM233073 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3786, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3786 | |
 | |
| KM233074 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3787, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3787 | |
 | |
| KM233075 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3788, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3788 | |
 | |
| KM233076 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3789.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3789.1 | |
 | |
| KM233077 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3795, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3795 | |
 | |
| KM233078 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3796, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3796 | |
 | |
| KM233079 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3798, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3798 | |
 | |
| KM233080 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3799, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3799 | |
 | |
| KM233081 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3800, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3800 | |
 | |
| KM233082 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.1 | |
 | |
| KM233083 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.2 | |
 | |
| KM233084 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3807, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3807 | |
 | |
| KM233085 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3808, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3808 | |
 | |
| KM233086 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3809, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3809 | |
 | |
| KM233087 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.1 | |
 | |
| KM233088 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.2 | |
 | |
| KM233089 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3814, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3814 | |
 | |
| KM233090 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3816, [coding-]complete genome2 = Zaire ebolavirus1 |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3816 | |
 | |
| KM233091 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3817, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3817 | |
 | |
| KM233092 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3818, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3818 | |
 | |
| KM233093 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3819, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3819 | |
 | |
| KM233094 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3820, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3820 | |
 | |
| KM233095 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3821, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3821 | |
 | |
| KM233096 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3822, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3822 | |
 | |
| KM233097 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3823, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3823 | |
 | |
| KM233098 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.1 | |
 | |
| KM233099 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.2 | |
 | |
| KM233100 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3826, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3826 | |
 | |
| KM233101 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3827, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3827 | |
 | |
| KM233102 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3829, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3829 | |
 | |
| KM233103 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3831, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona- G3831 | |
 | |
| KM233104 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3834, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3834 | |
 | |
| KM233105 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3838, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3838 | |
 | |
| KM233106 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3840, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3840 | |
 | |
| KM233107 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3841, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3841 | |
 | |
| KM233108 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3845, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3845 | |
 | |
| KM233109 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3846, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3846 | |
 | |
| KM233110 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3848, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3848 | |
 | |
| KM233111 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3850, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3850 | |
 | |
| KM233112 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3851, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3851 | |
 | |
| KM233113 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.1 | |
 | |
| KM233114 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.3 | |
 | |
| KM233115 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3857, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3857 | |
 | |
| KM233116 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.1 | |
 | |
| KM233117 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Mak-NM042.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.2 | |
 | |
| KM233118 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.3 | |
 | |
| KM251803 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-frag/NGA/2014/Makona-01072014 L gene, partial CDS3 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| <strain> = [EMPTY] | |
| </isolate> = Ebola virus/H.sapiens-frag/NGA/2014/Makona-01072014 | |
 |
4. The 2014 Ebola Virus Variant Originating in the Democratic Republic of the Congo


| GenBank Accession Number | <GenBank field> = Updated/corrected information |
|---|---|
| Final names | |
| KM517570 | <DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C15, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-frag/COD/2014/Lomela-Lokolia16 | |
 | |
| KM517571 | <DEFINITION LINE> = Zaire ebolavirus1 Ebola isolate virus H.sapiens-frag/COD/2014/Lomela-Lokolia17 L gene, partial CDS2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-frag/COD/2014/Lomela-Lokolia17 | |
 | |
| KM519951 | <DEFINITION LINE> = Zaire ebolavirus1 Ebola isolate virus H.sapiens-wt/COD/2014/Lomela-Lokolia1, [coding-]complete genome3 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/COD/2014/Lomela-Lokolia1 | |
 |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baize, S.; Pannetier, D.; Oestereich, L.; Rieger, T.; Koivogui, L.; Magassouba, N.; Soropogui, B.; Sow, M.S.; Keita, S.; De Clerck, H.; et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014, 371, 1418–1425. [Google Scholar] [PubMed]
- Kuhn, J.H.; Becker, S.; Ebihara, H.; Geisbert, T.W.; Jahrling, P.B.; Kawaoka, Y.; Netesov, S.V.; Nichol, S.T.; Peters, C.J.; Volchkov, V.E.; et al. Family Filoviridae. In Virus Taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier/Academic Press: London, UK, 2011; pp. 665–671. [Google Scholar]
- Calvignac-Spencer, S.; Schulze, J.M.; Zickmann, F.; Renard, B.Y. Clock Rooting Further Demonstrates that Guinea 2014 EBOV is a Member of the Zaire Lineage. PLoS Curr. 2014, 6. [Google Scholar] [CrossRef]
- Dudas, G.; Rambaut, A. Phylogenetic Analysis of Guinea 2014 EBOV Ebolavirus Outbreak. PLoS Curr. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Global Alert and Response (GAR). Situation Reports: Ebola Response Roadmap; World Health Organization: Geneva, Switzerland, 2014. Available online: http://apps.who.int/iris/bitstream/10665/136508/1/roadmapsitrep15Oct2014.pdf?ua=1 (accessed on 23 November 2014).
- Gire, S.K.; Goba, A.; Andersen, K.G.; Sealfon, R.S.; Park, D.J.; Kanneh, L.; Jalloh, S.; Momoh, M.; Fullah, M.; Dudas, G.; et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 2014, 345, 1369–1372. [Google Scholar] [CrossRef]
- Ladner, J.T.; Beitzel, B.; Chain, P.S.; Davenport, M.G.; Donaldson, E.F.; Frieman, M.; Kugelman, J.R.; Kuhn, J.H.; O’Rear, J.; Sabeti, P.C.; et al. Standards for sequencing viral genomes in the era of high-throughput sequencing. MBio 2014, 5, e01360–14. [Google Scholar] [PubMed]
- Kuhn, J.H. Filoviruses. A Compendium of 40 Years of Epidemiological, Clinical, and Laboratory Studies. Archives of Virology Supplementum; SpringerWienNewYork: Vienna, Austria, 2008; Volume 20. [Google Scholar]
- Maganga, G.D.; Kapetshi, J.; Berthet, N.; Ilunga, B.K.; Kabange, F.; Kingebeni, P.M.; Mondonge, V.; Muyembe, J.J.; Bertherat, E.; Briand, S.; et al. Ebola Virus Disease in the Democratic Republic of Congo. N. Engl. J. Med. 2014. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Bao, Y.; Bavari, S.; Becker, S.; Bradfute, S.; Brister, J.R.; Bukreyev, A.A.; Chandran, K.; Davey, R.A.; Dolnik, O.; et al. Virus nomenclature below the species level: A standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 2013, 158, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Andersen, K.G.; Bao, Y.; Bavari, S.; Becker, S.; Bennett, R.S.; Bergman, N.H.; Blinkova, O.; Bradfute, S.; Brister, J.R.; et al. Filovirus RefSeq Entries: Evaluation and Selection of Filovirus Type Variants, Type Sequences, and Names. Viruses 2014, 6, 3663–3682. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Becker, S.; Ebihara, H.; Geisbert, T.W.; Johnson, K.M.; Kawaoka, Y.; Lipkin, W.I.; Negredo, A.I.; Netesov, S.V.; Nichol, S.T.; et al. Proposal for a revised taxonomy of the family Filoviridae: Classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010, 155, 2083–2103. [Google Scholar] [CrossRef]
- Adams, M.J.; Carstens, E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch. Virol. 2012, 157, 1411–1422. [Google Scholar] [PubMed]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Carstens, E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2014). Arch. Virol. 2014, 159, 2831–2841. [Google Scholar] [CrossRef]
- Bukreyev, A.A.; Chandran, K.; Dolnik, O.; Dye, J.M.; Ebihara, H.; Leroy, E.M.; Muhlberger, E.; Netesov, S.V.; Patterson, J.L.; Paweska, J.T.; et al. Discussions and decisions of the 2012–2014 International Committee on Taxonomy of Viruses (ICTV) Filoviridae Study Group, January 2012-June 2013. Arch. Virol. 2014, 159, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Lofts, L.L.; Kugelman, J.R.; Smither, S.J.; Lever, M.S.; van der Groen, G.; Johnson, K.M.; Radoshitzky, S.R.; Bavari, S.; Jahrling, P.B.; et al. Reidentification of Ebola virus E718 and ME as Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Ecran. Genome Announc. 2014, 2, e01178–14. [Google Scholar] [PubMed]
- Schieffelin, J.S.; Shaffer, J.G.; Goba, A.; Gbakie, M.; Gire, S.K.; Colubri, A.; Sealfon, R.S.; Kanneh, L.; Moigboi, A.; Momoh, M.; et al. Clinical Illness and Outcomes in Patients with Ebola in Sierra Leone. N. Engl. J. Med. 2014. [Google Scholar] [CrossRef]
- Kreuels, B.; Wichmann, D.; Emmerich, P.; Schmidt-Chanasit, J.; de Heer, G.; Kluge, S.; Sow, A.; Renne, T.; Gunther, S.; Lohse, A.W.; et al. A Case of Severe Ebola Virus Infection Complicated by Gram-Negative Septicemia. N. Engl. J. Med. 2014. [Google Scholar] [CrossRef]
- Briand, S.; Bertherat, E.; Cox, P.; Formenty, P.; Kieny, M.P.; Myhre, J.K.; Roth, C.; Shindo, N.; Dye, C. The international Ebola emergency. N. Engl. J. Med. 2014, 371, 1180–1183. [Google Scholar] [CrossRef] [PubMed]
- Bah, E.I.; Lamah, M.C.; Fletcher, T.; Jacob, S.T.; Brett-Major, D.M.; Sall, A.A.; Shindo, N.; Fischer, W.A.; Lamontagne, F.; Saliou, S.M.; et al. Clinical Presentation of Patients with Ebola Virus Disease in Conakry, Guinea. N. Engl. J. Med. 2014. [Google Scholar] [CrossRef]
- Lyon, G.M.; Mehta, A.K.; Varkey, J.B.; Brantly, K.; Plyler, L.; McElroy, A.K.; Kraft, C.S.; Towner, J.S.; Spiropoulou, C.; Stroher, U.; et al. Clinical Care of Two Patients with Ebola Virus Disease in the United States. N. Engl. J. Med. 2014. [Google Scholar] [CrossRef]
- Barry, M.; Traore, F.A.; Sako, F.B.; Kpamy, D.O.; Bah, E.I.; Poncin, M.; Keita, S.; Cisse, M.; Toure, A. Ebola outbreak in Conakry, Guinea: Epidemiological, clinical, and outcome features. Med. Mal. Infect. 2014. [Google Scholar] [CrossRef]
- International Organization for Standardization (ISO) Country Codes—ISO 3166. Available online: https://www.iso.org/obp/ui/#search (accessed on 24 October 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuhn, J.H.; Andersen, K.G.; Baize, S.; Bào, Y.; Bavari, S.; Berthet, N.; Blinkova, O.; Brister, J.R.; Clawson, A.N.; Fair, J.; et al. Nomenclature- and Database-Compatible Names for the Two Ebola Virus Variants that Emerged in Guinea and the Democratic Republic of the Congo in 2014. Viruses 2014, 6, 4760-4799. https://doi.org/10.3390/v6114760
Kuhn JH, Andersen KG, Baize S, Bào Y, Bavari S, Berthet N, Blinkova O, Brister JR, Clawson AN, Fair J, et al. Nomenclature- and Database-Compatible Names for the Two Ebola Virus Variants that Emerged in Guinea and the Democratic Republic of the Congo in 2014. Viruses. 2014; 6(11):4760-4799. https://doi.org/10.3390/v6114760
Chicago/Turabian StyleKuhn, Jens H., Kristian G. Andersen, Sylvain Baize, Yīmíng Bào, Sina Bavari, Nicolas Berthet, Olga Blinkova, J. Rodney Brister, Anna N. Clawson, Joseph Fair, and et al. 2014. "Nomenclature- and Database-Compatible Names for the Two Ebola Virus Variants that Emerged in Guinea and the Democratic Republic of the Congo in 2014" Viruses 6, no. 11: 4760-4799. https://doi.org/10.3390/v6114760
APA StyleKuhn, J. H., Andersen, K. G., Baize, S., Bào, Y., Bavari, S., Berthet, N., Blinkova, O., Brister, J. R., Clawson, A. N., Fair, J., Gabriel, M., Garry, R. F., Gire, S. K., Goba, A., Gonzalez, J.-P., Günther, S., Happi, C. T., Jahrling, P. B., Kapetshi, J., ... Formenty, P. (2014). Nomenclature- and Database-Compatible Names for the Two Ebola Virus Variants that Emerged in Guinea and the Democratic Republic of the Congo in 2014. Viruses, 6(11), 4760-4799. https://doi.org/10.3390/v6114760