HIV-1 Replication and the Cellular Eukaryotic Translation Apparatus
Abstract
:1. Introduction
2. Overview of Eukaryotic Translation
2.1 Translation Initiation
2.1.1. Pre-Initiation Complex Assembly and mRNA Activation

2.1.2. Initiation Codon Recognition and 80S Complex Formation
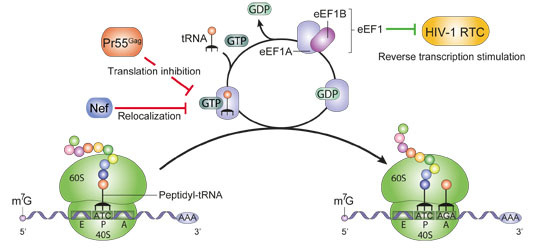
2.2. Translation Elongation and Termination
3. HIV-1 Takes Advantage of the Host Translation Machinery
3.1. Overcoming Ribosome Scanning Barriers

3.2. The Vpu-Env Bicistronic mRNA: Modulation of Ribosome Scanning Process
3.3. The Fate of Unspliced HIV-1 mRNA
3.3.1. Cap- and IRES-Dependent Translation Initiation


3.3.2. Ribosomal Frameshift

3.4. Redirecting mRNA Activation
3.5. Targeting Cellular Translation Factors
3.6. Inhibiting APOBEC3G Translation
4. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Izaurralde, E.; Lewis, J.; McGuigan, C.; Jankowska, M.; Darzynkiewicz, E.; Mattaj, I.W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 1994, 78, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.L.; Wente, S.R. Uncovering nuclear pore complexity with innovation. Cell 2013, 152, 1218–1221. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Proudfoot, N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009, 136, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Maquat, L.E.; Tarn, W.Y.; Isken, O. The pioneer round of translation: Features and functions. Cell 2010, 142, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Hoshino, S.I.; Imataka, H.; Sonenberg, N.; Katada, T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 2002, 277, 50286–50292. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Bandyopadhyay, A.; Maitra, U. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40S preinitiation complex. J. Biol. Chem. 2003, 278, 6580–6587. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature 1984, 308, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Kozak, C.A.; Chakraborti, A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 1996, 225, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Maag, D.; Fekete, C.A.; Gryczynski, Z.; Lorsch, J.R. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell 2005, 17, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Nanda, J.S.; Cheung, Y.N.; Takacs, J.E.; Martin-Marcos, P.; Saini, A.K.; Hinnebusch, A.G.; Lorsch, J.R. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 2009, 394, 268–285. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Lomakin, I.B.; Lee, J.H.; Choi, S.K.; Dever, T.E.; Hellen, C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 2000, 403, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Groppo, R.; Richter, J.D. Translational control from head to tail. Curr. Opin. Cell Biol. 2009, 21, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Kapp, L.D.; Lorsch, J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004, 73, 657–704. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 2012, 86, 45–93. [Google Scholar] [PubMed]
- Pisarev, A.V.; Skabkin, M.A.; Pisareva, V.P.; Skabkina, O.V.; Rakotondrafara, A.M.; Hentze, M.W.; Hellen, C.U.T.; Pestova, T.V. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 2010, 37, 196–210. [Google Scholar] [CrossRef] [PubMed]
- De Breyne, S.; Soto-Rifo, R.; López-Lastra, M.; Ohlmann, T. Translation initiation is driven by different mechanisms on the HIV-1 and HIV-2 genomic RNAs. Virus Res. 2013, 171, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Soto-Rifo, R.; Rubilar, P.S.; Limousin, T.; de Breyne, S.; Décimo, D.; Ohlmann, T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012, 31, 3745–3756. [Google Scholar] [CrossRef] [PubMed]
- Soto-Rifo, R.; Rubilar, P.S.; Ohlmann, T. The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res. 2013, 41, 6286–6299. [Google Scholar] [CrossRef] [PubMed]
- Groom, H.C.T.; Anderson, E.C.; Dangerfield, J.A.; Lever, A.M.L. Rev regulates translation of human immunodeficiency virus type 1 RNAs. J. Gen. Virol. 2009, 90, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Greatorex, J.; Gallego, J.; Varani, G.; Lever, A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J. Mol. Biol. 2002, 322, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Wang, S.W.; Cheng, L.; Tarn, W.Y.; Tsai, S.J.; Sun, H.S. Human DDX3 Interacts with the HIV-1 Tat Protein to Facilitate Viral mRNA Translation. PLoS One 2013, 8, e68665. [Google Scholar] [CrossRef] [PubMed]
- Fujii, R.; Okamoto, M.; Aratani, S.; Oishi, T.; Ohshima, T.; Taira, K.; Baba, M.; Fukamizu, A.; Nakajima, T. A Role of RNA Helicase A in cis-Acting Transactivation Response Element-mediated Transcriptional Regulation of Human Immunodeficiency Virus Type 1. J. Biol. Chem. 2001, 276, 5445–5451. [Google Scholar] [CrossRef] [PubMed]
- Dorin, D.; Bonnet, M.C.; Bannwarth, S.; Gatignol, A.; Meurs, E.F.; Vaquero, C. The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J. Biol. Chem. 2003, 278, 4440–4448. [Google Scholar] [CrossRef] [PubMed]
- Dugré-Brisson, S.; Elvira, G.; Boulay, K.; Chatel-Chaix, L.; Mouland, A.J.; DesGroseillers, L. Interaction of Staufen1 with the 5' end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005, 33, 4797–4812. [Google Scholar] [CrossRef] [PubMed]
- Svitkin, Y.V.; Pause, A.; Sonenberg, N. La autoantigen alleviates translational repression by the 5' leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994, 68, 7001–7007. [Google Scholar] [PubMed]
- Schwartz, S.; Felber, B.K.; Pavlakis, G.N. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol. Cell. Biol. 1992, 12, 207–219. [Google Scholar] [PubMed]
- Schwartz, S.; Felber, B.K.; Fenyö, E.M.; Pavlakis, G.N. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 1990, 64, 5448–5456. [Google Scholar] [PubMed]
- Anderson, J.L.; Johnson, A.T.; Howard, J.L.; Purcell, D.F.J. Both linear and discontinuous ribosome scanning are used for translation initiation from bicistronic human immunodeficiency virus type 1 env mRNAs. J. Virol. 2007, 81, 4664–4676. [Google Scholar] [CrossRef] [PubMed]
- Krummheuer, J.; Johnson, A.T.; Hauber, I.; Kammler, S.; Anderson, J.L.; Hauber, J.; Purcell, D.F.J.; Schaal, H. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology 2007, 363, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Brasey, A.; Lopez-Lastra, M.; Ohlmann, T.; Beerens, N.; Berkhout, B.; Darlix, J.L.; Sonenberg, N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003, 77, 3939–3949. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Arts, K.; Abbink, T.E.M. Ribosomal scanning on the 5'-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 2011, 39, 5232–5244. [Google Scholar] [CrossRef] [PubMed]
- Monette, A.; Valiente-Echeverría, F.; Rivero, M.; Cohen, E.A.; Lopez-Lastra, M.; Mouland, A.J. Dual Mechanisms of Translation Initiation of the Full-Length HIV-1 mRNA Contribute to Gag Synthesis. PloS One 2013, 8, e68108. [Google Scholar] [CrossRef] [PubMed]
- Ricci, E.P.; Soto Rifo, R.; Herbreteau, C.H.; Decimo, D.; Ohlmann, T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem. Soc. Trans. 2008, 36, 690–693. [Google Scholar] [CrossRef] [PubMed]
- Miele, G.; Mouland, A.; Harrison, G.P.; Cohen, E.; Lever, A.M. The human immunodeficiency virus type 1 5' packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996, 70, 944–951. [Google Scholar] [PubMed]
- Buck, C.B.; Shen, X.; Egan, M.A.; Pierson, T.C.; Walker, C.M.; Siliciano, R.F. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001, 75, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Gendron, K.; Ferbeyre, G.; Heveker, N.; Brakier-Gingras, L. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 2011, 39, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, M.; Carvajal, F.; Pino, K.; Navarrete, C.; Ferres, M.; Huidobro-Toro, J.P.; Sargueil, B.; López-Lastra, M. Functional and Structural Analysis of the Internal Ribosome Entry Site Present in the mRNA of Natural Variants of the HIV-1. PLoS One 2012, 7, e35031. [Google Scholar] [CrossRef] [PubMed]
- Amorim, R.; Costa, S.M.; Cavaleiro, N.P.; da Silva, E.E.; da Costa, L.J. HIV-1 transcripts use IRES-initiation under conditions where Cap-dependent translation is restricted by poliovirus 2A protease. PloS One 2014, 9, e88619. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, M.; Deforges, J.; Plank, T.D.M.; Letelier, A.; Ramdohr, P.; Abraham, C.G.; Valiente-Echeverría, F.; Kieft, J.S.; Sargueil, B.; López-Lastra, M. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 2011, 39, 6186–6200. [Google Scholar] [CrossRef] [PubMed]
- Plank, T.D.M.; Whitehurst, J.T.; Kieft, J.S. Cell type specificity and structural determinants of IRES activity from the 5' leaders of different HIV-1 transcripts. Nucleic Acids Res. 2013, 41, 6698–6714. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Henao-Mejia, J.; Liu, H.; Zhao, Y.; He, J.J. Translational Regulation of HIV-1 Replication by HIV-1 Rev Cellular Cofactors Sam68, eIF5A, hRIP, and DDX3. J. Neuroimmune Pharmacol. 2011, 6, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Monette, A.; Ajamian, L.; López-Lastra, M.; Mouland, A.J. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: Implications for HIV-1 gene expression. J. Biol. Chem. 2009, 284, 31350–31362. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Aravena, A.; Ramdohr, P.; Vallejos, M.; Valiente-Echeverría, F.; Dormoy-Raclet, V.; Rodríguez, F.; Pino, K.; Holzmann, C.; Huidobro-Toro, J.P.; Gallouzi, I.E.; et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology 2009, 392, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Valiente-Echeverría, F.; Vallejos, M.; Monette, A.; Pino, K.; Letelier, A.; Huidobro-Toro, J.P.; Mouland, A.J.; López-Lastra, M. A cis-acting element present within the Gag open reading frame negatively impacts on the activity of the HIV-1 IRES. PloS One 2013, 8, e56962. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yoder, K.; Hansen, M.S.; Olvera, J.; Miller, M.D.; Bushman, F.D. Retroviral cDNA integration: Stimulation by HMG I family proteins. J. Virol. 2000, 74, 10965–10974. [Google Scholar] [CrossRef] [PubMed]
- Holloway, A.F.; Occhiodoro, F.; Mittler, G.; Meisterernst, M.; Shannon, M.F. Functional interaction between the HIV transactivator Tat and the transcriptional coactivator PC4 in T cells. J. Biol. Chem. 2000, 275, 21668–21677. [Google Scholar] [CrossRef] [PubMed]
- Weill, L.; James, L.; Ulryck, N.; Chamond, N.; Herbreteau, C.H.; Ohlmann, T.; Sargueil, B. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010, 38, 1367–1381. [Google Scholar] [CrossRef] [PubMed]
- Brierley, I.; Dos Ramos, F.J. Programmed ribosomal frameshifting in HIV-1 and the SARS–CoV. Virus Res. 2006, 119, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Giedroc, D.P.; Cornish, P.V. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res. 2009, 139, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Staple, D.W.; Butcher, S.E. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005, 349, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yang, Y.; Wang, G.; Cheng, Q.; Du, Z. Highly conserved RNA pseudoknots at the Gag-Pol junction of HIV-1 suggest a novel mechanism of −1 ribosomal frameshifting. RNA N. Y. N 2014, 20, 587–593. [Google Scholar] [CrossRef]
- Wang, G.; Yang, Y.; Huang, X.; Du, Z. Possible involvement of coaxially stacked double pseudoknots in the regulation of −1 programmed ribosomal frameshifting in RNA viruses. J. Biomol. Struct. Dyn. 2014, 1–11. [Google Scholar]
- Low, J.T.; Garcia-Miranda, P.; Mouzakis, K.D.; Gorelick, R.J.; Butcher, S.E.; Weeks, K.M. Structure and Dynamics of the HIV-1 Frameshift Element RNA. Biochemistry (Mosc.) 2014, 53, 4282–4291. [Google Scholar] [CrossRef]
- Mouzakis, K.D.; Lang, A.L.; Meulen, K.A.V.; Easterday, P.D.; Butcher, S.E. HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome. Nucleic Acids Res. 2012. [Google Scholar] [CrossRef]
- Léger, M.; Dulude, D.; Steinberg, S.V.; Brakier-Gingras, L. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed −1 ribosomal frameshift. Nucleic Acids Res. 2007, 35, 5581–5592. [Google Scholar] [CrossRef] [PubMed]
- Namy, O.; Moran, S.J.; Stuart, D.I.; Gilbert, R.J.C.; Brierley, I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 2006, 441, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.Y.; Choi, Y.S.; Dinman, J.D.; Lee, K.H. The many paths to frameshifting: Kinetic modelling and analysis of the effects of different elongation steps on programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2011, 39, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Gendron, K.; Charbonneau, J.; Dulude, D.; Heveker, N.; Ferbeyre, G.; Brakier-Gingras, L. The presence of the TAR RNA structure alters the programmed −1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008, 36, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, J.; Gendron, K.; Ferbeyre, G.; Brakier-Gingras, L. The 5'UTR of HIV-1 full-length mRNA and the Tat viral protein modulate the programmed −1 ribosomal frameshift that generates HIV-1 enzymes. RNA N. Y. N 2012, 18, 519–529. [Google Scholar] [CrossRef]
- Lorgeoux, R.P.; Pan, Q.; Le Duff, Y.; Liang, C. DDX17 promotes the production of infectious HIV-1 particles through modulating viral RNA packaging and translation frameshift. Virology 2013, 443, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Kronja, I.; Orr-Weaver, T.L. Translational regulation of the cell cycle: When, where, how and why? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2011, 366, 3638–3652. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005, 433, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Planelles, V.; Barker, E. Roles of Vpr and Vpx in modulating the virus-host cell relationship. Mol. Asp. Med. 2010, 31, 398–406. [Google Scholar] [CrossRef]
- Sharma, A.; Yilmaz, A.; Marsh, K.; Cochrane, A.; Boris-Lawrie, K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012, 8, e1002612. [Google Scholar] [CrossRef] [PubMed]
- Ventoso, I.; Blanco, R.; Perales, C.; Carrasco, L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl. Acad. Sci. USA 2001, 98, 12966–12971. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, E.; Castelló, A.; Menéndez-Arias, L.; Carrasco, L. HIV protease cleaves poly(A)-binding protein. Biochem. J. 2006, 396, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Castelló, A.; Franco, D.; Moral-López, P.; Berlanga, J.J.; Alvarez, E.; Wimmer, E.; Carrasco, L. HIV-1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PloS One 2009, 4, e7997. [Google Scholar] [CrossRef] [PubMed]
- Ohlmann, T.; Prévôt, D.; Décimo, D.; Roux, F.; Garin, J.; Morley, S.J.; Darlix, J.L. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002, 318, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV-human protein complexes. Nature 2012, 481, 365–370. [Google Scholar]
- Clerzius, G.; Gélinas, J.F.; Gatignol, A. Multiple levels of PKR inhibition during HIV-1 replication. Rev. Med. Virol. 2011, 21, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Sadler, A.J.; Williams, B.R.G. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 2007, 316, 253–292. [Google Scholar] [PubMed]
- Mcmillan, N.A.J.; Chun, R.F.; Siderovski, D.P.; Galabru, J.; Toone, W.M.; Samuel, C.E.; Mak, T.W.; Hovanessian, A.G.; Jeang, K.T.; Williams, B.R.G. HIV-1 Tat Directly Interacts with the Interferon-Induced, Double-Stranded RNA-Dependent Kinase, PKR. Virology 1995, 213, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, P.A.; Anderson, E.; Lary, J.; Cole, J.L. Mechanism of PKR Activation by dsRNA. J. Mol. Biol. 2008, 381, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Clerzius, G.; Shaw, E.; Daher, A.; Burugu, S.; Gélinas, J.F.; Ear, T.; Sinck, L.; Routy, J.P.; Mouland, A.J.; Patel, R.C.; Gatignol, A. The PKR activator, PACT, becomes a PKR inhibitor during HIV-1 replication. Retrovirology 2013, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Cimarelli, A.; Luban, J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1999, 73, 5388–5401. [Google Scholar] [PubMed]
- Khan, R.; Giedroc, D.P. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J. Biol. Chem. 1992, 267, 6689–6695. [Google Scholar] [PubMed]
- Merrick, W.C. Eukaryotic Protein Synthesis: Still a Mystery. J. Biol. Chem. 2010, 285, 21197–21201. [Google Scholar] [CrossRef] [PubMed]
- Ott, D.E.; Coren, L.V.; Johnson, D.G.; Kane, B.P.; Sowder, R.C., 2nd; Kim, Y.D.; Fisher, R.J.; Zhou, X.Z.; Lu, K.P.; Henderson, L.E. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 2000, 266, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wei, T.; Abbott, C.M.; Harrich, D. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol. Mol. Biol. Rev. MMBR 2013, 77, 253–266. [Google Scholar] [CrossRef]
- Abbas, W.; Khan, K.A.; Tripathy, M.K.; Dichamp, I.; Keita, M.; Rohr, O.; Herbein, G. Inhibition of ER stress-mediated apoptosis in macrophages by nuclear-cytoplasmic relocalization of eEF1A by the HIV-1 Nef protein. Cell Death Dis. 2012, 3, e292. [Google Scholar] [CrossRef] [PubMed]
- Warren, K.; Wei, T.; Li, D.; Qin, F.; Warrilow, D.; Lin, M.H.; Sivakumaran, H.; Apolloni, A.; Abbott, C.M.; Jones, A.; Anderson, J.L.; Harrich, D. Eukaryotic elongation factor 1 complex subunits are critical HIV-1 reverse transcription cofactors. Proc. Natl. Acad. Sci. USA 2012, 109, 9587–9592. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Henriet, S.; Mercenne, G.; Bernacchi, S.; Paillart, J.C.; Marquet, R. Tumultuous Relationship between the Human Immunodeficiency Virus Type 1 Viral Infectivity Factor (Vif) and the Human APOBEC-3G and APOBEC-3F Restriction Factors. Microbiol. Mol. Biol. Rev. 2009, 73, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Mercenne, G.; Bernacchi, S.; Richer, D.; Bec, G.; Henriet, S.; Paillart, J.C.; Marquet, R. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010, 38, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Mariani, R.; Chen, D.; Schröfelbauer, B.; Navarro, F.; König, R.; Bollman, B.; Münk, C.; Nymark-McMahon, H.; Landau, N.R. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 2003, 114, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.; Khan, M.A.; Miyagi, E.; Plishka, R.; Buckler-White, A.; Strebel, K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 2003, 77, 11398–11407. [Google Scholar] [CrossRef] [PubMed]
- Stopak, K.; de Noronha, C.; Yonemoto, W.; Greene, W.C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 2003, 12, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Batisse, J.; Guerrero, S.; Bernacchi, S.; Sleiman, D.; Gabus, C.; Darlix, J.L.; Marquet, R.; Tisné, C.; Paillart, J.C. The role of Vif oligomerization and RNA chaperone activity in HIV-1 replication. Virus Res. 2012, 169, 361–376. [Google Scholar] [CrossRef] [PubMed]
- Sleiman, D.; Bernacchi, S.; Xavier Guerrero, S.; Brachet, F.; Larue, V.; Paillart, J.C.; Tisne, C. Characterization of RNA binding and chaperoning activities of HIV-1 Vif protein: Importance of the C-terminal unstructured tail. RNA Biol. 2014, 11, 906–920. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T. Ribosome profiling: New views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014, 15, 205–213. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Zarnack, K.; Rot, G.; Curk, T.; Kayikci, M.; Zupan, B.; Turner, D.J.; Luscombe, N.M.; Ule, J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010, 17, 909–915. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerrero, S.; Batisse, J.; Libre, C.; Bernacchi, S.; Marquet, R.; Paillart, J.-C. HIV-1 Replication and the Cellular Eukaryotic Translation Apparatus. Viruses 2015, 7, 199-218. https://doi.org/10.3390/v7010199
Guerrero S, Batisse J, Libre C, Bernacchi S, Marquet R, Paillart J-C. HIV-1 Replication and the Cellular Eukaryotic Translation Apparatus. Viruses. 2015; 7(1):199-218. https://doi.org/10.3390/v7010199
Chicago/Turabian StyleGuerrero, Santiago, Julien Batisse, Camille Libre, Serena Bernacchi, Roland Marquet, and Jean-Christophe Paillart. 2015. "HIV-1 Replication and the Cellular Eukaryotic Translation Apparatus" Viruses 7, no. 1: 199-218. https://doi.org/10.3390/v7010199
APA StyleGuerrero, S., Batisse, J., Libre, C., Bernacchi, S., Marquet, R., & Paillart, J. -C. (2015). HIV-1 Replication and the Cellular Eukaryotic Translation Apparatus. Viruses, 7(1), 199-218. https://doi.org/10.3390/v7010199