Field Efficacy and Transmission of Fast- and Slow-Killing Nucleopolyhedroviruses that Are Infectious to Adoxophyes honmai (Lepidoptera: Tortricidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Viruses
2.3. Killing Speed of AdhoNPV and AdorNPV
2.4. Field Cage Trials

2.5. Position of Infected Insect Cadavers
2.6. Statistical Analysis
3. Results and Discussion
3.1. Killing Speed

3.2. Field Efficacy within Sprayed Populations


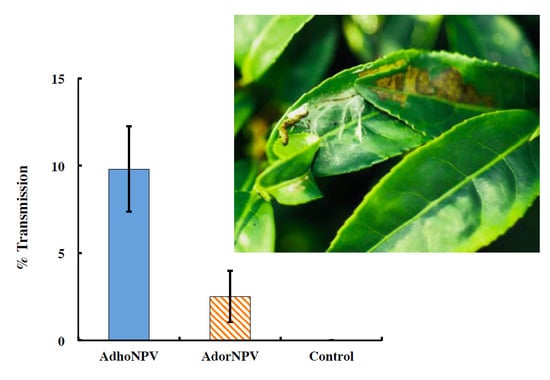
3.3. Viral Transmission between Generations


4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tamaki, Y. Tortircids in tea. In Tortricid Pests, Their Biology, Natural Enemies and Control; Van der Geest, L.P.S., Evenhuis, H.H., Eds.; Elsevier: Amsterdam, The Netherland, 1991; pp. 541–551. [Google Scholar]
- Szewczyk, B.; Hoyos-Carvajal, L.; Paluszek, M.; Skrzecz, W.; de Souza, M.L. Baculoviruses—Re-emerging biopesticides. Biotechnol. Adv. 2006, 24, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Cory, J.S.; Myers, J.H. The ecology and evolution of insect baculoviruses. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 239–272. [Google Scholar] [CrossRef]
- Yamada, H.; Oho, N. A granulosis virus, possible biological agent for control of Adoxophyes orana (Lepidoptera: Tortricidae) in apple orchards: I. Mass production. J. Invertebr. Pathol. 1973, 21, 144–148. [Google Scholar] [CrossRef]
- Sato, T.; Oho, N.; Kodomari, S. A granulosis virus of the tea tortrix, Homon magnanima DIAKNOFF (Lepidoptera: Tortricidae); Its pathogenicity and Mass-Production method. Appl. Entomol. Zool. 1980, 15, 409–415. [Google Scholar]
- Asser-Kaiser, S.; Fritsch, E.; Undorf-Spahn, K.; Kienzle, J.; Eberle, K.E.; Gund, N.A.; Reineke, A.; Zebitz, C.P.W.; Heckel, D.G.; Huber, J.; et al. Rapid emergence of baculovirus resistance in codling moth due to dominant, sex-linked inheritance. Science 2007, 317, 1916–1918. [Google Scholar]
- Gebhardt, M.M.; Eberle, K.E.; Radtke, P.; Jehle, J.A. Baculovirus resistance in codling moth is virus isolate-dependent and the consequence of a mutation in viral gene pe38. Proc. Natl. Acad. Sci. USA 2014, 111, 15711–15716. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M.; Takeda, M.; Kunimi, Y. Seasonal changes in prevalence of viral disease and parasitism by parasitic insects in a larval population of the smaller tea tortrix, Adoxophyes sp. (Lepidoptera: Tortricidae) in a tea field. Appl. Entomol. Zool. 1997, 32, 609–615. [Google Scholar]
- Ishii, T.; Nakai, M.; Okuno, S.; Takatsuka, J.; Kunimi, Y. Characterization of Adoxophyeshonmai single-nucleocapsid nucleopolyhedrovirus: Morphology, structure, and effects on larvae. J. Invertebr. Pathol. 2003, 83, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, J.; Okuno, S.; Ishii, T.; Nakai, M.; Kunimi, Y. Fitness-related traits of entomopoxviruses isolated from Adoxophyes honmai (Lepidoptera: Tortricidae) at three locations in Japan. J. Invertebr. Pathol. 2010, 105, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hilton, S.; Winstanley, D. Genomic sequence and biological characterization of a nucleopolyhedrovirus isolated from the summer fruit tortrix, Adoxophyes orana. J. Gen. Virol. 2008, 89, 2898–2908. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Nakai, M.; Nakanishi, K.; Sato, T.; Hilton, S.; Winstanley, D.; Kunimi, Y. Genetic and biological comparisons of four nucleopolyhedrovirus isolates that are infectious to Adoxophyes honmai (Lepidoptera: Tortricidae). Biol. Control 2008, 46, 542–546. [Google Scholar] [CrossRef]
- Nakai, M.; Goto, C.; Kang, W.; Shikata, M.; Luque, T.; Kunimi, Y. Genome sequence and organization of a nucleopolyhedrovirus isolated from the smaller tea tortrix, Adoxophyes honmai. Virology 2003, 316, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Nabeta, F.H.; Nakai, M.; Kunimi, Y. Effects of temperature and photoperiod on the development and reproduction of Adoxophyes honmai (Lepidoptera: Tortricidae). Appl. Entomol. Zool. 2005, 40, 231–238. [Google Scholar] [CrossRef]
- Harrison, R.L. Genomic sequence analysis of the Illinois strain of the Agrotis ipsilon multiple nucleopolyhedrovirus. Virus Genes 2009, 38, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Bonning, B.C.; Hammock, B.D. Development of recombinant baculoviruses for insect control. Annu. Rev. Entomol. 1996, 41, 191–210. [Google Scholar] [CrossRef] [PubMed]
- Bonning, B.C.; Boughton, A.J.; Jin, H.L.; Harrison, R.L. Genetic enhancement of baculovirus insecticides. In Advances in Microbial Control of Insect Pests; Upadhyay, R.K., Ed.; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2003; pp. 109–126. [Google Scholar]
- Cory, J.S.; Hirst, M.L.; Williams, T.; Hails, R.S.; Goulson, D.; Green, B.M.; Carty, T.M.; Possee, R.D.; Cayley, P.J.; Bishop, D.H.L.; et al. Field trial of a genetically improved baculovirus insecticide. Nature 1994, 370, 138–140. [Google Scholar]
- Hoover, K.; Schultz, C.M.; Lane, S.S.; Bonning, B.C.; Duffey, S.S.; Mccutchen, B.F.; Hammock, B.D. Reduction in damage to cotton plants by a recombinant baculovirus that knocks moribund larvae of Heliothis virescens off the plant. Biol. Control 1995, 5, 419–426. [Google Scholar] [CrossRef]
- Inceoglu, A.B.; Kamita, S.G.; Hammock, B.D. Genetically modified baculoviruses: A historical overview and future outlook. Adv. Virus Res. 2006, 68, 323–360. [Google Scholar] [PubMed]
- Anderson, R.M.; May, R.M. The population dynamics of microparasites and their invertebrate hosts. Philos. Trans. R. Soc. B Biol. Sci. 1981, 291, 451–524. [Google Scholar] [CrossRef]
- Hochberg, M.E. The potential role of pathogens in biological control. Nature 1989, 337, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D. Wipfelkrankheit: Modification of host behavior during baculoviral infection. Oecologia 1997, 109, 219–228. [Google Scholar] [CrossRef]
- Hoover, K.; Grove, M.; Gardner, M.; Hughes, D.P.; McNeil, J.; Slavicek, J. A gene for an extended phenotype. Science 2011, 333, 1401. [Google Scholar] [CrossRef] [PubMed]
- Katsuma, S.; Koyano, Y.; Kang, W.; Kokusho, R.; Kamita, S.G.; Shimada, T. The baculovirus uses a captured host phosphatase to induce enhanced locomotory activity in host caterpillars. PLoS Pathog. 2012, 8, e1002644. [Google Scholar] [CrossRef] [PubMed]
- Kamita, S.G.; Nagasaka, K.; Chua, J.W.; Shimada, T.; Mita, K.; Kobayashi, M.; Maeda, S.; Hammock, B.D. A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc. Natl. Acad. Sci. USA 2005, 102, 2584–2589. [Google Scholar] [CrossRef] [PubMed]
- Van Houte, S.; Ros, V.I.D.; Mastenbroek, T.G.; Vendrig, N.J.; Hoover, K.; Spitzen, J.; van Oers, M.M. Protein tyrosine phosphatase-induced hyperactivity is a conserved strategy of a subset of baculoviruses to manipulate lepidopteran host behavior. PLoS One 2012, 7, e46933. [Google Scholar] [CrossRef] [PubMed]
- Ros, V.I.D.; van Houte, S.; Hemerik, L.; van Oers, M.M. Baculovirus-induced tree-top disease: How extended is the role of egt as a gene for the extended phenotype? Mol. Ecol. 2014, 28. [Google Scholar] [CrossRef]
- Lee, Y.; Fuxa, J.R.; Inceoglu, A.B.; Alaniz, S.A.; Richter, A.R.; Reilly, L.M.; Hammock, B.D. Competition between wild-type and recombinant nucleopolyhedroviruses in a greenhouse microcosm. Biol. Control 2001, 20, 84–93. [Google Scholar] [CrossRef]
- Burand, J.P.; Nakai, M.; Smith, I. Host-Virus Interactions. In Insect Pathogens, Molecular Approaches and Techniques; Stock, S.P., Vandenberg, J., Glazer, I., Boemare, N., Eds.; CAB International: Oxfordshire, UK, 2009; pp. 195–222. [Google Scholar]
- Hawtin, R.E.; Zarkowska, T.; Arnold, K.; Thomas, C.J.; Gooday, G.W.; King, L.A.; Kuzio, J.A.; Possee, R.D. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 1997, 238, 243–253. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, V.; Slavicek, J.; Podgwaite, J.D.; Webb, R.; Fuester, R.; Peiffer, R.A. Deletion of v-chiA from a baculovirus reduces horizontal transmission in the field. Appl. Environ. Microbiol. 2013, 79, 4056–4064. [Google Scholar] [CrossRef] [PubMed]
- Fuxa, J.R. Ecology of insect nucleopolyhedroviruses. Agric. Ecosyst. Environ. 2004, 103, 27–43. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, M.; Nakai, M.; Saito, Y.; Sato, Y.; Ishijima, C.; Kunimi, Y. Field Efficacy and Transmission of Fast- and Slow-Killing Nucleopolyhedroviruses that Are Infectious to Adoxophyes honmai (Lepidoptera: Tortricidae). Viruses 2015, 7, 1271-1283. https://doi.org/10.3390/v7031271
Takahashi M, Nakai M, Saito Y, Sato Y, Ishijima C, Kunimi Y. Field Efficacy and Transmission of Fast- and Slow-Killing Nucleopolyhedroviruses that Are Infectious to Adoxophyes honmai (Lepidoptera: Tortricidae). Viruses. 2015; 7(3):1271-1283. https://doi.org/10.3390/v7031271
Chicago/Turabian StyleTakahashi, Maho, Madoka Nakai, Yasumasa Saito, Yasushi Sato, Chikara Ishijima, and Yasuhisa Kunimi. 2015. "Field Efficacy and Transmission of Fast- and Slow-Killing Nucleopolyhedroviruses that Are Infectious to Adoxophyes honmai (Lepidoptera: Tortricidae)" Viruses 7, no. 3: 1271-1283. https://doi.org/10.3390/v7031271
APA StyleTakahashi, M., Nakai, M., Saito, Y., Sato, Y., Ishijima, C., & Kunimi, Y. (2015). Field Efficacy and Transmission of Fast- and Slow-Killing Nucleopolyhedroviruses that Are Infectious to Adoxophyes honmai (Lepidoptera: Tortricidae). Viruses, 7(3), 1271-1283. https://doi.org/10.3390/v7031271