The Effect of MicroRNA bantam on Baculovirus AcMNPV Infection in Vitro and in Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, MicroRNA Mimic, Inhibitor and AntagomiR
2.2. Transfection and Infection
2.3. Real-Time PCR
2.4. Insect Experiments
3. Results
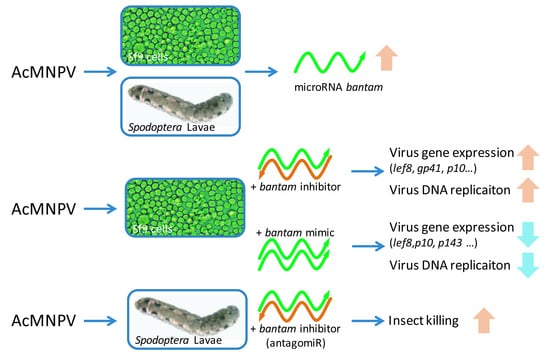
3.1. bantam Level after Infection
3.2. The Effect of bantam Mimic and Inhibitor on AcMNPV Replication in Sf9 Cells
3.3. The Effect of bantam antagomiR in AcMNPV Infection in Spodoptera larvae
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MOI | multiplicity of infection |
| AcMNPV | Autographa californica multicapsid nucleopolyhedrovirus |
References
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. MicroRNAs as mediators of viral evasion of the immune system. Nat. Immunol. 2013, 14, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.E.; Steitz, J.A. Virus meets host microRNA: The destroyer, the booster, the hijacker. Mol. Cell. Biol. 2014, 34, 3780–3787. [Google Scholar] [CrossRef] [PubMed]
- Buggele, W.A.; Johnson, K.E.; Horvath, C.M. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J. Biol. Chem. 2012, 287, 31027–31040. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, N.J.; Hayward, S.L.; Crowe, J.E., Jr. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB. MBio 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Backes, S.; Shapiro, J.S.; Sabin, L.R.; Pham, A.M.; Reyes, I.; Moss, B.; Cherry, S.; tenOever, B.R. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 2012, 12, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Jeang, K.-T. RNAi in the regulation of mammalian viral infections. BMC Biol. 2012, 10. [Google Scholar] [CrossRef] [PubMed]
- Sanghvi, V.R.; Steel, L.F. RNA silencing as a cellular defense against HIV-1 infection: Progress and issues. FASEB J. 2012, 26, 3937–3945. [Google Scholar] [CrossRef] [PubMed]
- Nathans, R.; Chu, C.-Y.; Serquina, A.K.; Lu, C.-C.; Cao, H.; Rana, T.M. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 2009, 34, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.K.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of Hepatitis C virus RNA abundance by a liver-specific microRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Moosmann, A.; Groemminger, S.; Walz, N.; Grundhoff, A.; Hammerschmidt, W. MicroRNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010, 6, e1001063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feederle, R.; Linnstaedt, S.D.; Bannert, H.; Lips, H.; Bencun, M.; Cullen, B.R.; Delecluse, H.-J. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011, 7, e1001294. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xu, H.; Yao, Y.; Smith, L.P.; Kgosana, L.; Green, J.; Petherbridge, L.; Baigent, S.J.; Nair, V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 2011, 7, e1001305. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, M.; Hussain, M.; Asgari, S. MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. J. Gen. Virol. 2013, 94, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.J.; Cohen, S.M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 2006, 126, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Becam, I.; Rafel, N.; Hong, X.; Cohen, S.M.; Milan, M. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development 2011, 138, 3781–3789. [Google Scholar] [CrossRef] [PubMed]
- Herranz, H.; Hong, X.; Cohen, S.M. Mutual repression by bantam miRNA and capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol. 2012, 22, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Harti, M.; Kadow, I.C.G. New roles for “old” microRNAs in nervous system function and disease. Front. Mol. Neurosci. 2013, 6. [Google Scholar] [CrossRef]
- Dong, L.; Li, J.; Huang, H.; Yin, M.-X.; Xu, J.; Li, P.; Lu, Y.; Wu, W.; Yang, H.; Zhao, Y.; et al. Growth suppressor lingerer regulates bantam microRNA to restrict organ size. J. Mol. Cell Biol. 2015, 7, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Bras-Pereira, C.; Casares, F.; Janody, F. The retinal determination gene dachshund restricts cell proliferation by limiting the activity of the homothorax-yorkie complex. Development 2015, 142, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, D.; Pflugfelder, G.O.; Shen, J. Dpp signaling inhibits proliferation in the Drosophila wing by Omb-dependent regional control of bantam. Development 2013, 140, 2917–2922. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Padgett, R.W. bantam Is required for optic lobe development and glial cell proliferation. PLoS ONE 2012, 7, e32910. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Boulan, L.; Martin, D.; Milan, M. bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr. Biol. 2013, 23, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Lerner, I.; Bartok, O.; Wolfson, V.; Menet, J.S.; Weissbein, U.; Afik, S.; Haimovich, D.; Gafni, C.; Friedman, N.; Rosbash, M.; et al. Clk post-transcriptional control denoises circadian transcription both temporally and spatially. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, L.; Ding, S.; Huang, X.; Zhang, J.; Yin, J.; Zhong, J. Chicken HS4 insulator significantly improves baculovirus-mediated foreign gene expression in insect cells by modifying the structure of neighbouring chromatin in virus minichromosome. J. Biotechnol. 2009, 142, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Kingsolver, M.B.; Huang, Z.; Hardy, R.W. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar] [CrossRef] [PubMed]
- Zografidis, A.; van Nieuwerburgh, F.; Kolliopoulou, A.; Apostolou-Karampelis, K.; Head, S.R.; Deforce, D.; Smagghe, G.; Swevers, L. Viral small-RNA analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. J. Virol. 2015, 89, 11473–11486. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Smagghe, G.; de Coninck, D.I.; van Nieuwerburgh, F.; Deforce, D.; Meeus, I. In vivo study of Dicer-2-mediated immune response of the small interfering RNA pathway upon systemic infections of virulent and avirulent viruses in Bombus terrestris. Insect Biochem. Mol. Biol. 2016, 70, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Asgari, S. MicroRNAs as mediators of insect host-pathogen interactions and immunity. J. Insect Physiol. 2014, 70, 151–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Gao, S.; Zhang, D.; Yin, J.; Xiang, Z.; Xia, Q. MicroRNAs show diverse and dynamic expression patterns in multiple tissues of Bombyx mori. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zuo, H.; Liu, K.; Li, H.; Zhong, G. A new strategy for identification of highly conserved microRNAs in non-model insect, Spodoptera litura. Int. J. Mol. Sci. 2012, 13, 612–627. [Google Scholar] [CrossRef] [PubMed]
- Etebari, K.; Hussain, M.; Asgari, S. Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem. Mol. Biol. 2013, 43, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Sun, J.; Zhang, Q.; Wang, Z.; Huang, Y.; Pan, W. Identification and characterization of novel microRNAs from Schistosoma japonicum. PLoS ONE 2008, 3, e4034. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhao, J.; Wang, J.; Hu, C.; Peng, J.; Luo, R.; Zhou, C.; Liu, J.; Lin, J.; Jin, Y.; et al. MicroRNAs are involved in the regulation of ovary development in the pathogenic blood fluke Schistosoma japonicum. PLoS Pathog. 2016, 12, e1005423. [Google Scholar] [CrossRef] [PubMed]
- Nahvi, A.; Shoemaker, C.J.; Green, R. An expanded seed sequence definition accounts for full regulation of the hid 3′ UTR by bantam miRNA. RNA 2009, 15, 814–822. [Google Scholar] [CrossRef] [PubMed]







| Target | Type | Sequence (5′–3′) |
|---|---|---|
| bantam | primer assay | CTTTCTGAGATCATTGTGAAAG |
| miR-750 | primer assay | CAGTTGCCAGATCTATCTTTC |
| U6 (B. mori) | primer assay | AGAGACGATTAGCATGGCCC |
| ie-0 | upper primer | CATATTCGTGCGAGGCAACG |
| lower primer | CGAGTTGACGCTTGCCAAAA | |
| ie-1 | upper primer | TTTTAACGCGTCGTACACCA |
| lower primer | GTTGACGCTTGCCAAAAAGT | |
| ie2 | upper primer | AGCGAAGAAAACGTGCAGAT |
| lower primer | CTCCGACGCAATGTTATCCT | |
| lef-1 | upper primer | TTCCGCATATGCAAGATTCA |
| lower primer | ACCACATCCACCAGTTCCAT | |
| lef-3 | upper primer | AACCCCCATGAGCGACTATTTG |
| lower primer | ACCGCTGTTGATTTTCTTGTA | |
| lef-8 | upper primer | AATGGCTACTCTGTGGCGGTAA |
| lower primer | TTACCGCCACAGAGTAGCCATT | |
| egt | upper primer | ACCGTTTCCAGCGATCAACT |
| lower primer | TAGAGGCGGAAACGTTGCTT | |
| me53 | upper primer | CGCATCTCTCCGCCTAAACA |
| lower primer | GCGGGCAAACCTTCAAACTT | |
| p143 | upper primer | AAGTATTTGCCCGAGGAC |
| lower primer | ATTTTGGCGATGTGATAAGT | |
| p35 | upper primer | ACGACACGGGACTTTACGAG |
| lower primer | GTTTTTCGACGCTTCGTTGT | |
| vlf-1 | upper primer | CGTGCCAATTCCAACGGTTT |
| lower primer | CAACTCAGCGTGGACGATCT | |
| gp41 | upper primer | AGAGTTGGGACAGAGCAACG |
| lower primer | GCGCCACCGTTGTAAAACTT | |
| orf-1629 | upper primer | GTTAGGCACGGGAGAA |
| lower primer | CGAAGCAGACGACCTT | |
| p10 | upper primer | GACGCCGTTACGGAAACTAA |
| lower primer | GTCTGGAAGATCCGGAACAA | |
| egfp | upper primer | CGTAAACGGCCACAAGTTCAT |
| lower primer | GGGTCAGCTTGCCGTAGGT | |
| orf-75 | upper primer | GCTGGCAGTTGGTATGCTTC |
| lower primer | ACTCAGTAGGCGACAGGTTG | |
| β-tublin (B. mori) | upper primer | TTGCATTGGTACACTGGCGA |
| lower primer | ACACCAGGTCGTTCATGTTGC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Ran, Z.; Li, S.; Yin, J.; Zhong, J. The Effect of MicroRNA bantam on Baculovirus AcMNPV Infection in Vitro and in Vivo. Viruses 2016, 8, 136. https://doi.org/10.3390/v8050136
Shi X, Ran Z, Li S, Yin J, Zhong J. The Effect of MicroRNA bantam on Baculovirus AcMNPV Infection in Vitro and in Vivo. Viruses. 2016; 8(5):136. https://doi.org/10.3390/v8050136
Chicago/Turabian StyleShi, Xiaojie, Zihan Ran, Sisi Li, Juan Yin, and Jiang Zhong. 2016. "The Effect of MicroRNA bantam on Baculovirus AcMNPV Infection in Vitro and in Vivo" Viruses 8, no. 5: 136. https://doi.org/10.3390/v8050136
APA StyleShi, X., Ran, Z., Li, S., Yin, J., & Zhong, J. (2016). The Effect of MicroRNA bantam on Baculovirus AcMNPV Infection in Vitro and in Vivo. Viruses, 8(5), 136. https://doi.org/10.3390/v8050136