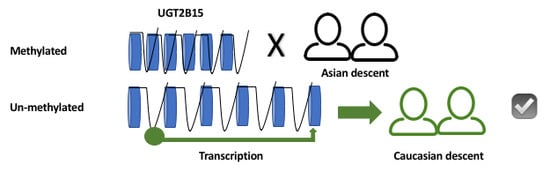
Regulation of Hepatic UGT2B15 by Methylation in Adults of Asian Descent
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Availability and Collection
2.2. Liver Samples
2.3. DNA and RNA Extraction
2.4. DNA Methylation Analysis
2.5. Gene Expression via Quantitative Reverse Transcriptase Polymerase Chain Reaction (q-RT-PCR)
2.6. Genotyping of the UGT2B17 Deletion
2.7. UGT2B15 Polymerase Chain Reaction (PCR) Amplification for Promoter Region
2.8. UGT2B15 PCR Amplification for 3′ Untranslated Regulatory Region
2.9. Cycle Sequencing
2.10. Statistical Analyses
3. Results
3.1. Summary of Methylation Beta Values for All UGT-Associated Loci
3.2. The mRNA Expression of UDP-Glucuronosyl Transferase (UGT) Isoforms in This Liver Cohort
3.3. UGT2B15 and UGT2B17 Genetic Analysis
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Miners, J.O.; Mackenzie, P.I. Drug glucuronidation in humans. Pharmacol. Ther. 1991, 51, 347–369. [Google Scholar] [CrossRef]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Bock, K.W.; Burchell, B.; Guillemette, C.; Ikushiro, S.; Iyanagi, T.; Miners, J.O.; Owens, I.S.; Nebert, D.W. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genom. 2005, 15, 677–685. [Google Scholar] [CrossRef]
- Miners, J.O.; Smith, P.A.; Sorich, M.J.; McKinnon, R.A.; Mackenzie, P.I. Predicting human drug glucuronidation parameters: Application of in vitro and in silico modeling approaches. Ann. Rev. Pharmacol. Toxicol. 2004, 44, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Izukawa, T.; Nakajima, M.; Fujiwara, R.; Yamanaka, H.; Fukami, T.; Takamiya, M.; Aoki, Y.; Ikushiro, S.; Sakaki, T.; Yokoi, T. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab. Dispos. 2009, 37, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Koszik, F.; Brunner, P.; Stingl, G. “Overrepresentation of T17 cells in the peripheral blood of psoriatic patients is not confined to the skin-homing T cell subset”. J. Dermatol. Sci. 2014, 75, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Belanger, A.S.; Tojcic, J.; Harvey, M.; Guillemette, C. Regulation of UGT1A1 and HNF1 transcription factor gene expression by DNA methylation in colon cancer cells. BMC Mol. Biol. 2010, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, J.F.; Bernard, O.; Villeneuve, L.; Tetu, B.; Guillemette, C. Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer. Clin. Cancer Res. 2006, 12, 1850–1858. [Google Scholar] [CrossRef] [PubMed]
- Yasar, U.; Greenblatt, D.J.; Guillemette, C.; Court, M.H. Evidence for regulation of UDP-glucuronosyltransferase (UGT) 1A1 protein expression and activity via DNA methylation in healthy human livers. J. Pharm. Pharmacol. 2013, 65, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W., 3rd.; Pardo-Manuel de Villena, F.; Lyn-Cook, B.D.; Chatterjee, P.K.; Bell, T.A.; Detwiler, D.A.; Gilmore, R.C.; Valladeras, I.C.; Wright, C.C.; Threadgill, D.W.; et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics 2004, 84, 707–714. [Google Scholar] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-blast: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Trottier, J.; Verreault, M.; Grepper, S.; Monte, D.; Belanger, J.; Kaeding, J.; Caron, P.; Inaba, T.T.; Barbier, O. Human UDP-glucuronosyltransferase (UGT) 1A3 enzyme conjugates chenodeoxycholic acid in the liver. Hepatology 2006, 44, 1158–1170. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.Z.; Cox, L.S.; Ahluwalia, J.S.; Renner, C.C.; Hatsukami, D.K.; Benowitz, N.L.; Tyndale, R.F. Genetic and phenotypic variation in UGT2B17, a testosterone-metabolizing enzyme, is associated with BMI in males. Pharmacogenet. Genom. 2015, 25, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Gutknecht, N.; van Gogswaardt, D.; Conrads, G.; Apel, C.; Schubert, C.; Lampert, F. Diode laser radiation and its bactericidal effect in root canal wall dentin. J. Clin. Laser Med. Surg. 2000, 18, 57–60. [Google Scholar] [PubMed]
- Wagner, J.R.; Busche, S.; Ge, B.; Kwan, T.; Pastinen, T.; Blanchette, M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014, 15, R37. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Court, M.H.; Duan, S.X.; Guillemette, C.; Journault, K.; Krishnaswamy, S.; Von Moltke, L.L.; Greenblatt, D.J. Stereoselective conjugation of oxazepam by human UDP-glucuronosyltransferases (UGTs): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9. Drug Metab. Dispos. 2002, 30, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Hesse, L.M.; Hazarika, S.; Masse, G.; Harmatz, J.S.; Greenblatt, D.J.; Court, M.H. Evidence for oxazepam as an in vivo probe of UGT2B15: Oxazepam clearance is reduced by UGT2B15 D85Y polymorphism but unaffected by UGT2B17 deletion. Br. J. Clin. Pharmacol. 2009, 68, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Chen, G.; Dellinger, R.W.; Sharma, A.K.; Lazarus, P. Characterization of 17-dihydroexemestane glucuronidation: Potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet. Genom. 2010, 20, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Trucksis, M.; McElwee, J.J.; Wong, P.H.; Maciolek, C.; Thompson, C.D.; Prueksaritanont, T.; Garrett, G.C.; Declercq, R.; Vets, E.; et al. Ugt2B17 genetic polymorphisms dramatically affect the pharmacokinetics of MK-7246 in healthy subjects in a first-in-human study. Clin. Pharmacol. Ther. 2012, 92, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Stingl, J.C.; Bartels, H.; Viviani, R.; Lehmann, M.L.; Brockmoller, J. Relevance of UDP-glucuronosyltransferase polymorphisms for drug dosing: A quantitative systematic review. Pharmacol. Ther. 2014, 141, 92–116. [Google Scholar] [CrossRef] [PubMed]
- Belanger, A.; Pelletier, G.; Labrie, F.; Barbier, O.; Chouinard, S. Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol. Metab. 2003, 14, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, G.; Bellemare, J.; Audet-Walsh, E.; Flageole, C.; Huang, S.P.; Bao, B.Y.; Douville, P.; Caron, P.; Fradet, Y.; Lacombe, L.; et al. Deletions of the androgen-metabolizing UGT2B genes have an effect on circulating steroid levels and biochemical recurrence after radical prostatectomy in localized prostate cancer. J. Clin. Endocrinol. Metab. 2011, 96, E1550–E1557. [Google Scholar] [CrossRef] [PubMed]
- Sten, T.; Finel, M.; Ask, B.; Rane, A.; Ekstrom, L. Non-steroidal anti-inflammatory drugs interact with testosterone glucuronidation. Steroids 2009, 74, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Sten, T.; Bichlmaier, I.; Kuuranne, T.; Leinonen, A.; Yli-Kauhaluoma, J.; Finel, M. Udp-glucuronosyltransferases (UGTs) 2B7 and UGT2B17 display converse specificity in testosterone and epitestosterone glucuronidation, whereas UGT2A1 conjugates both androgens similarly. Drug Metab. Dispos. 2009, 37, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Swanson, C.; Mellstrom, D.; Lorentzon, M.; Vandenput, L.; Jakobsson, J.; Rane, A.; Karlsson, M.; Ljunggren, O.; Smith, U.; Eriksson, A.L.; et al. The uridine diphosphate glucuronosyltransferase 2B15 D85Y and 2B17 deletion polymorphisms predict the glucuronidation pattern of androgens and fat mass in men. J. Clin. Endocrinol. Metab. 2007, 92, 4878–4882. [Google Scholar] [CrossRef] [PubMed]
- Gauthier-Landry, L.; Belanger, A.; Barbier, O. Multiple roles for UDP-glucuronosyltransferase (UGT) 2B15 and UGT2B17 enzymes in androgen metabolism and prostate cancer evolution. J. Steroid Biochem. Mol. Biol. 2014, 145, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Kpoghomou, M.A.; Soatiana, J.E.; Kalembo, F.W.; Bishwajit, G.; Sheng, W. UGT2B17 polymorphism and risk of prostate cancer: A meta-analysis. ISRN Oncol. 2013, 201. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Chen, L.; Shade, K.; Lazarus, P.; Seigne, J.; Patterson, S.; Helal, M.; Pow-Sang, J. Asp85tyr polymorphism in the UDP-glucuronosyltransferase (UGT) 2B15 gene and the risk of prostate cancer. J. Urol. 2004, 171, 2484–2488. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, S.L.; Nowell, S.; Plaxco, J.; Lang, N.P. An allele-specific polymerase chain reaction method for the determination of the D85Y polymorphism in the human UDP-glucuronosyltransferase 2B15 gene in a case-control study of prostate cancer. Ann. Surg. Oncol. 2000, 7, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Whittemore, A.S.; Kolonel, L.N.; Wu, A.H.; John, E.M.; Gallagher, R.P.; Howe, G.R.; Burch, J.D.; Hankin, J.; Dreon, D.M.; West, D.W.; et al. Prostate cancer in relation to diet, physical activity, and body size in blacks, whites, and Asians in the United States and Canada. JNCI 1995, 87, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Kayser, M.; Brauer, S.; Weiss, G.; Underhill, P.A.; Roewer, L.; Schiefenhovel, W.; Stoneking, M. Melanesian origin of Polynesian Y chromosomes. Curr. Biol. 2000, 10, 1237–1246. [Google Scholar] [CrossRef]
- Kayser, M.; Brauer, S.; Cordaux, R.; Casto, A.; Lao, O.; Zhivotovsky, L.A.; Moyse-Faurie, C.; Rutledge, R.B.; Schiefenhoevel, W.; Gil, D.; et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol. Biol. Evol. 2006, 23, 2234–2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Y.; Meng, X.; Liu, J.; Yan, L.; Wang, P.; Bi, H.; Kan, Q.; Zhang, L. Histone modifications regulate the developmental expression of human hepatic UDP-glucuronosyltransferase 1A1. Drug Metab. Dispos. 2017, 45, 1372–1378. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, N.; Petroczi, A.; Barker, J.; Szekely, A.D.; Hussain, I.; Naughton, D.P. Potentially harmful advantage to athletes: A putative connection between UGT2B17 gene deletion polymorphism and renal disorders with prolonged use of anabolic androgenic steroids. Subst. Abuse Treat. Prev. Policy 2010, 5, 7. [Google Scholar] [CrossRef] [PubMed]




| UGT | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|
| 1A1 | AATAAAAAAGGACTCTGCTATGCT | ACATCAAAGCTGCTTTCTGC |
| 1A3 | TGTTGAACAATATGTCTTTGGTCTA | ACCACATCAAAGGAAGTAGCA |
| 1A4 | GAACAATGTATCTTTGGCCC | ACCACATCAAAGGAAGTAGCA |
| 1A6 | CATGATTGTTATTGGCCTGTAC | TCTGTGAAAAGAGCATCAAACT |
| 1A8 | GAAAGCACAAGTACGAAGTTTG | GGGAGGGAGAAATATTTGGC |
| 1A9 | TGGAAAGCACAAGTACGAAGTATATA | GGGAGGGAGAAATATTTGGC |
| 1A10 | GAAAGCACAGGCACAAAGTATA | GGGAGGGAGAAATATTTAGCAAC |
| 2B4 | TCTTTCGATCCAACAGCC | CATCTCTTAACCGCTGCTTGATA |
| 2B7 | GGAGAATTTCTCATGCAACAGA | CAGAACTTTCTATTATGTCACCAAATATTG |
| 2B11 | AGTAACATGACAGCAGAAAGGGCCAAT | AGACCTAAGGCATCTGGTTTATTCCCG |
| 2B15 | CTTCTGAAAATCTCGATAGATGGAT | CATCTTTACAGACTTGTTACTGTAGTCAT |
| 2B17 | TTTATGAAAAGTTCGATAGATGGAC | CATCTTCACAGACTTTATATTATAGTCAG |
| GUSB | AGCCAGTTCCTCATCAATGG | GGTAGTGGCTGGTACGGAAA |
| UGT | Region | Primer | Orientation | Sequence (5′-3′) | Amplicon (bp) |
|---|---|---|---|---|---|
| 2B17 | Gene | C | Forward | CCTGGAAGAGCTTGTTCAGA | 316 |
| 2B17 | Gene | C | Reverse | CTGCATCTTCACAGAGCTTT | 316 |
| 2B17 | Deletion | J | Forward | TGCACAGAGTTAAGAAATGGAGAGATGTG | 884 |
| 2B17 | Deletion | J | Reverse | GATCATCCTATATCCTGACAGAATTCTTTTG | 884 |
| 2B15 | Promoter | A | Forward | GGTCCCACTTCTTCAGATCAT | 3368 |
| 2B15 | Promoter | A | Reverse | GAGAGAAGGAAGAAGCCAGAAG | 3368 |
| 2B15 | Promoter | B | Forward | ACATAGGAAGGAGGGAACAGA | 3224 |
| 2B15 | Promoter | B | Reverse | TTCCTGCTGAGGGTTTGAAG | 3224 |
| 2B15 | UTR | D | Forward | TGGTGTGGATGTCCTTTCTG | 2567 |
| 2B15 | UTR | D | Reverse | GGCAGGAGAATGACTTGACTAC | 2567 |
| 2B15 | UTR | E | Forward | CTGCAGGTCTGTTGGAATTTG | 2401 |
| 2B15 | UTR | E | Reverse | GCAGTTGTAGTCCTAGCTTCTC | 2401 |
| UGT Isoform | Region | Primer # | Strand | Sequence (5′-3′) |
|---|---|---|---|---|
| 2B17 | Gene | 1 | − | CTGGTCCCACTTCTTCAGAT |
| 2B15 | Promoter | 2 | + | GTTTGCAGATTTTTAATGAGGCA |
| 2B15 | Promoter | 3 | + | CTCCTAGGATTTGGCACCAG |
| 2B15 | Promoter | 4 | + | TTCTCTAATTTGACTCAGCTTCACA |
| 2B15 | Promoter | 5 | − | CTCAGCCCACCTGCAACC |
| 2B15 | Promoter | 6 | − | CCCCCTCTCCAGAATACACA |
| 2B15 | Promoter | 7 | − | TATCGTGGTGCAAGTAATGTCTTC |
| 2B15 | Promoter | 8 | − | TTATCCAATGGCTGTATTCTGTG |
| 2B15 | Promoter | 9 | + | ACTTTCCCACCGAAAATTCC |
| 2B15 | Promoter | 10 | + | TGCGTGGCAACTGTGATATT |
| 2B15 | Promoter | 11 | − | CAGGAAAAAGGAAATCCTCCA |
| 2B15 | Promoter | 12 | − | CTTTCGTGTGTAACTTTTGGATT |
| 2B15 | UTR | 13 | + | GAGGTTACTGCTGTCTCTTTGT |
| 2B15 | UTR | 14 | + | TGGTGTGGATGTCCTTTCTG |
| 2B15 | UTR | 15 | − | CCCTGGATCGAGCAGTCTTC |
| 2B15 | UTR | 16 | − | GACCAACCAATGAAGCCCCT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oeser, S.G.; Bingham, J.-P.; Collier, A.C. Regulation of Hepatic UGT2B15 by Methylation in Adults of Asian Descent. Pharmaceutics 2018, 10, 6. https://doi.org/10.3390/pharmaceutics10010006
Oeser SG, Bingham J-P, Collier AC. Regulation of Hepatic UGT2B15 by Methylation in Adults of Asian Descent. Pharmaceutics. 2018; 10(1):6. https://doi.org/10.3390/pharmaceutics10010006
Chicago/Turabian StyleOeser, Steffen G., Jon-Paul Bingham, and Abby C. Collier. 2018. "Regulation of Hepatic UGT2B15 by Methylation in Adults of Asian Descent" Pharmaceutics 10, no. 1: 6. https://doi.org/10.3390/pharmaceutics10010006
APA StyleOeser, S. G., Bingham, J. -P., & Collier, A. C. (2018). Regulation of Hepatic UGT2B15 by Methylation in Adults of Asian Descent. Pharmaceutics, 10(1), 6. https://doi.org/10.3390/pharmaceutics10010006