Aptamers in Virology—A Consolidated Review of the Most Recent Advancements in Diagnosis and Therapy
Abstract
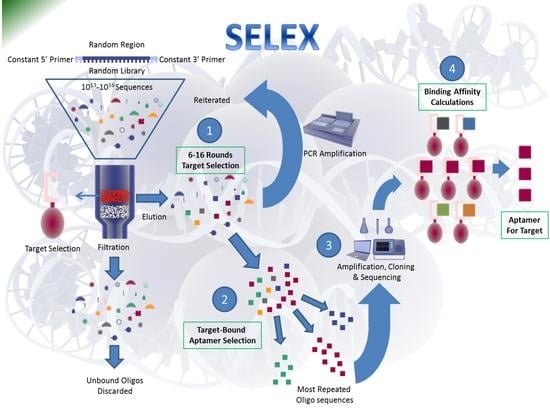
:1. Introduction
2. Human Immunodeficiency Virus (HIV)
2.1. HIV Structure and Entry
2.2. Aptamers in Anti-HIV Therapy
2.3. Aptamers in HIV Detection
3. Hepatitis Virus (HBV, HCV)
3.1. HBV Structure and Lifecycle
3.2. HCV Structure and Lifecycle
3.3. Aptamers for Hepatitis Virus Therapy
3.4. Aptamers for Hepatitis Virus Diagnosis
4. Influenza Virus
4.1. Structure and Lifecycle of Influenza-A Virus
4.2. Aptamers for the Treatment of the Influenza Virus
4.3. Aptamers for the Detection of the Influenza Virus
5. Herpes Simplex Virus (HSV)
5.1. HSV Structure and Lifecycle
5.2. Aptamers for HSV
6. Discussion
6.1. Role of Modern Technologies in Developing Aptamers
6.2. Role of Computational Approaches in Developing Aptamers
6.3. Aptamers against the SARS-CoV-2 Pandemic
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Reference | Detection/Therapy | Mode of Detection/Therapy | Detection Range or Binding Affinity | Target | RNA/DNA | Size | Year |
|---|---|---|---|---|---|---|---|
| [47] | Detection | - | low pM | HIV-1 RT | RNA | 20 + 57 nt | 2015 |
| [46] | Detection | - | 30 aM detection | - | - | - | 2015 |
| [45] | Detection | Atomic Force Microscopy | 0.8 nM to 8 pM | gp120 | DNA | 23 kDa | 2014 |
| [43] | Detection | rGO Flourescence | - | interferon-γ | DNA | - | 2014 |
| [44] | Detection | Kd = 2–4 nM | 28 nM | nucleocapsid protein 7 (NCp7) | RNA | 20 nt SL3 Aptamer | 2013 |
| [41] | Detection | Diamond-FET-based-sensing | 1 nM | HIV-1 Tat protein | RNA | 19 nt Sigma Aldrich, St. Louis, MO 68178, USA | 2013 |
| [42] | Detection | Imaging | Kd 2.5 × 10−8 M | HIV-1 RT | DNA | 93 nt G-Quadruplex Sangon Biotech, 698 Xiangyu Road, Songjiang District, Shanghai Zip Code: 201611 - | 2011 |
| [40] | Detection | Diamond-FET-based sensing | 1 pM to 1 μg/ml | HIV 1 tat peptide | RNA | 36 nt Sigma Genesis company, St. Louis, MO 68178, USA | 2011 |
| [39] | Detection and Therapy | Transfection-based assay | Prediction of best inhibitors | HIV RT | RNA | - | 2014 |
| [48] | Detection | MST Measurement | 10 µM | HIV-1 integrase | DNA | 2016 | |
| [49] | Detection | QIAamp kit | 29 nM | gp120 | RNA | 29 nt | 2016 |
| [50] | Detection | qPCR | gag protein | RNA | 80 nt | 2017 | |
| [50] | Detection | ELISA | High affinity | RT | RNA | 2017 | |
| [51] | Detection | SELEX | High affinity | RT | DNA | 31 nt | 2017 |
| [52] | Detection | G-quadruplex/QTAp | High affinity | Tat protein | RNA | 14 nt | 2017 |
| [53] | Therapy | AS1411 | Clinical Trials | Nucleolin | RNA | - | 2015 |
| [54] | Therapy | Anti-TNPO3 siRNA | - | anti-CCR5 receptor | RNA | - | 2015 |
| [17] | Therapy | Live cell SELEX | - | anti-CCR5 receptor | RNA | - | 2015 |
| [22] | Therapy | Protease inhibition | - | aspartyl protease | RNA | - | 2015 |
| [22] | Therapy | Entry Inhibitors | - | gp120, gp41 | RNA | - | 2015 |
| [19] | Therapy | Chimeras | - | gp120 or CD4 | RNA | - | 2015 |
| [21] | Therapy | Stops HIV-C from causing cardiomyopathy | - | UCLA1 | RNA | Modified UCLA | 2014 |
| [16] | Therapy | Aptamer shortening up to their minimal active domain | 85% inhibition of HIV | 5′-untranslated region of HIV-1 genome | RNA | 16 nt | 2014 |
| [20] | Therapy | EC50 4.9–10 µM | - | gp120 | DNA | G-quadruplex-forming d(TG3AG) | 2014 |
| [18] | Therapy | RNA screening system | - | HIV RT | RNA | - | 2014 |
| [58] | Therapy | 70% inhibition | Kd = 1.59 nM | CD40 | DNA | 45 nt | 2014 |
| [34] | Therapy | - | - | gp120, gp41 and HSA | DNA | G-quadruplex-forming d(TG3AG) | 2014 |
| [59] | Therapy | Functionalized Gold nano | 40.2% decreased infectivity | HIV-1 RT | RNA | IDT company, 1710 Commercial Park, Coralville, Iowa 52241, USA | 2013 |
| [60] | Therapy | Hydrophobic aptamer | - | gp41 N-terminal heptad repeat (NHR) | DNA | DNA Duplex | 2013 |
| [19] | Therapy | Reduced topical dosage compard to earlier studies | - | CD4 | RNA | 40 nt | 2013 |
| [38] | Therapy | - | IC50 10 nM | HIV-1 RT | RNA | 31 nt | 2013 |
| [61] | Therapy | Binding Inhibition | - | P24 antigen | RNA | - | 2013 |
| [62] | Therapy | Rev Aptamer improvement | - | arginine-rich motif (ARM) of HIV Rev protein | RNA | 30 nt | 2013 |
| [37] | Therapy | High Through-put sequencing enhances chances for better selection | - | HIV-1 RT | RNA | 70 nt | 2013 |
| [63] | Therapy | IC50 value of 0.5 mM | Kd = 29–381 nM | interleukin-6 | DNA | 60 nt | 2013 |
| [16] | Therapy | Chemical Modification to attach siRNA | - | gp120 | RNA | - | 2013 |
| [36] | Therapy | Intracellular Aptamer | 5–10 fold suppression | HIV RT | RNA | - | 2012 |
| [35] | Therapy | Structural Investigation of G-Quadruplexes | - | - | DNA | G-quadruplex-forming d(TG3AG) | 2012 |
| [64] | Therapy | siRNA attached to Aptamer | - | CD4 | DNA | Made from RNA Aptamer 39 nt | 2012 |
| [65] | Therapy | - | - | CCR5 | DNA | 23 nt | 2012 |
| [66] | Therapy | Blocking HIV translation | Kd = 1.28 ± 1.27 nmol/l | human cyclin T1 | RNA | 40 nt | 2012 |
| [67] | Therapy | - | Kd = 82 ± 7 nM | HIV RT | DNA | G-Quadruplex | 2012 |
| [20] | Therapy | IC50 0.8 ± 0.9 nM | Kd = 0.15 nM | gp120 | RNA | 54 nt UCLA-1 first report | 2012 |
| [68] | Therapy | Stops Early intracellular events | - | Thrombin, RT | DNA | 3 DNA G-Quadruplexes | 2011 |
| [69] | Therapy | Targeted Delievery of pRNA | Kd = 47.91 nM | gp120 | RNA | 81 nt Aptamer | 2011 |
| [18] | Therapy | RNA aptamer with siRNA Chimera | - | CD4 | RNA | 86 nt-Aptamer A1 | 2011 |
| [70] | Therapy | Aptamer-siRNA Chimera | Kd = 47.91 nM | gp120 | RNA | 86 nt-Aptamer A1 | 2011 |
| [71] | Therapy | HSCs are engineered to express anti-HIV molecules | Kd = 80 to 200 nM | gag polyprotein | RNA | 100 nt | 2011 |
| Reference | Detection/Therapy | Mode of Detection/Therapy | Detection Range or Binding Affinity | Target | RNA/DNA | Size | Year |
|---|---|---|---|---|---|---|---|
| [90] | Detection - HBV | Chemiluminiscence | 0.5ng/L | Surface Antigen | DNA | - | 2015 |
| [89] | Detection-HBV | one-step competitive binding assay | 1.25 mIU mL-1 | surface antigen of the hepatitis B virus | RNA | 99 nt | 2014 |
| [94] | Detection-HCV | ELISA | - | glycoproteins, E1 and E2 | RNA | 2015 | |
| [92,93] | Detection-HCV | ELASA-First time | 0.8–4 nM | HCV E2 | DNA | 40 nt | 2013 |
| [92] | Detection -HCV | lateral flow strip | 10 pg/mL | HCV core antigen | DNA | - | 2013 |
| [95] | Detection-HCV | Malachite green flourescence | 250 nM | hepatitis C helicase and replicase | RNA | - | 2013 |
| [91] | Detection-HCV | Octet interferometer | 700 pg/ml | NS5B viral protein | RNA | 21 nt NS5B RNA BIONEER | 2011 |
| [188] | Detection-HBV | SELEX | 4 µM | HbeAg | DNA | 40 nt | 2016 |
| [206] | Detection-HBV | AgNC and MoS2 nanosheets | 10.7 nM | pLDH | DNA | 29 nt | 2017 |
| [195] | Detection-HCV | nanoaptasensor | 10 µm | HCVcoreAg | DNA | 2017 | |
| [214] | Detection-HCV | ELISA | 10−10 M | HCVcoreAg | DNA | 85 nt | 2018 |
| [211] | Detection-HCV | High affinity | Ribozyme | RNA | 2017 | ||
| [204] | Detection-HCV | ELISA | HCV polyprotein | DNA | 80 nt | 2017 | |
| [78,79] | Therapy-HBV | inhibition | 180 ± 82 nM | Surface protein L | DNA | - | 2015 |
| [78] | Therapy-HBV | inhibit the assembly of the nucleocapsid | - | core protein of HBV | DNA | Sangon Biotech, 698 Xiangyu Road, Songjiang District, Shanghai Zip Code: 201611 | 2014 |
| [76] | Therapy-HBV | Inhibition | - | HBV P protein | RNA | 29 nt | 2011 |
| [96] | Therapy-HDV | Detection of HDV Riboswitch | ON/OFF 4.7 | HDV Ribozyme | RNA | 8 nt | 2013 |
| [85,88] | Therapy-HCV | Chol attached Aptamer | - | nonstructural protein 5B | RNA | 29 nt | 2015 |
| [88,97] | Therapy-HCV | Magnetic Separation | - | E1E2 glycoprotein | RNA | - | 2015 |
| [97] | Therapy-HCV | inhibited HCV RNA replication | - | NS2 protein | DNA | 40 nt | 2014 |
| [98] | Therapy-HCV | Inhibition | - | core protein | DNA | 40 nt | 2014 |
| [99] | Therapy-HCV | Inhibition | - | NS5A | DNA | 40 nt | 2014 |
| [87] | Therapy-HCV | blockage of virus binding to cells | EC50 62.37 nM | envelope protein (E1E2) | DNA | 40 nt | 2013 |
| [84] | Therapy-HCV | competitive sequestration of the target protein | 1–3 nM | HCV NS5B RNA replicas | RNA | 29 nt | 2013 |
| [80,82] | Therapy-HCV | interference of HCV replication | 0.4–0.5 μM | CRE-5BSL3.2 domain | RNA | 30 nt | 2013 |
| [80] | Therapy-HCV | - | 80% inhibition of Viral RNA | HCV-CRE194 RNA fragments | RNA | 30 nt | 2012 |
| Reference | Influenza Type | Detection/Therapy | Mode of Detection/Therapy | Detection Range or Binding Affinity | Target | RNA/DNA | Size | Year |
|---|---|---|---|---|---|---|---|---|
| [136] | Detection | - | 1 μg/mL | viral nucleoprotein and vasopressin | DNA | Nucleoprotein - 63 nt AptaRes Ag & Vasopressin-54 nt IDT company, 1710 Commercial Park, Coralville, Iowa 52241, USA | 2012 | |
| [137] | influenza A/H1N1 | Detection | Microfluidic SELEX | Kd = 55.14 ± 22.40 nM | Whole Virus | DNA | 40 nt | 2014 |
| [138] | Both A and B | Detection | Gold nano attachment to virus | Kd = 44 ± 6 nM | hemagglutinin (HA) and neuraminidase (NA) | RNA | Various sizes | 2014 |
| [119] | H1N1 | Detection | ELISA/SPR | nM range | Surface protein hemagglutinin HA1 subunit of subtype H1 | DNA | G-quadruplex | 2015 |
| [139] | H3N2, H2N2, H5N1, H1N1 | Detection | Kd Tokio 0.7 ± 0.2 and Jilin 1.2 ± 0.2 μg/mL | Tokio virus 8 ng/mL Jilin-HA 800 ng/mL | Viral SELEX | RNA | 25 nt | 2013 |
| [133] | H5N1 | Detection | Sandwich assay amperometric | 100 fM | H5N1 specific | DNA | 72 nt | 2015 |
| [131] | H5N1 | Detection | Impedance | 0.0128 hemagglutinin units (HAU) | H5N1 specific | DNA | - | 2015 |
| [130] | H5N1 | Detection | Gold Nanogate enzymatic sensor | 2–9 HAU | H5N1 | DNA | - | 2015 |
| [140] | H5N1 | Detection | metal-enhanced fluorescence (MEF) sensing platform | 2 and 3.5 ng/mL | recombinant hemagglutinin (rHA) protein | DNA | G-quadruplex | 2015 |
| [129] | H5N1 | Detection | enzymatic catalysis with electrochemical impedance | 8 × 10−4 HAU in 200 μL | recombinant IGF-I | DNA | 72-nt biotin labelled | 2014 |
| [128] | H5N1 | Detection | (Kd) of 4.65nM | 12.8 HAU | hemagglutinin | DNA | N74 | 2013 |
| [127] | H5N1 | Detection | QCM based sensor | 0.0128 to 128 HAU | H5N1 surface protein | DNA | 75 nt IDT Company, 1710 Commercial Park, Coralville, Iowa 52241, USA | 2013 |
| [126] | H5N1 | Detection | SPR based portable sensor | 0.128 to 1.28 HAU | haemagglutinin (HA | DNA | 74 nt | 2012 |
| [132] | H5N1 | Detection | Electrochemical | 4.3 × 10−13 M/L | AIV H5N1 gene sequences | DNA | 23 nt Takara Biotech, 2560 Orchard Parkway, San Jose, CA 95131, USA | 2011 |
| [141] | H5N1, H1N1, and H3N2 | Detection | sandwich enzyme-linked aptamer assay | Kd 1.53 × 10−8 M | HA1 protein | DNA | - | 2014 |
| [142] | Influenza A | Detection | Labelling | Visual | hemagglutinin (HA) | DNA | 68 nt Sangon biotech, 698 Xiangyu Road, Songjiang District, Shanghai Zip Code: 201611 | 2011 |
| [135] | - | Detection | SERS | 0.1 μg/ml | Viral Nucleoprotein | DNA | 22 nt AptaRes AG | 2011 |
| [103] | Influenza A | Detection | activatable silver nanoclusters beacon | High affinity | H1N1/H5N1 genes | DNA | small nt sequence | 2017 |
| [143] | H3N2 | Detection | Impedimetric glycan-based biosensor | 5 aM | Glycan | DNA | 2016 | |
| [144] | H5N1 | Detection | nanowell-based QCM aptasensor | 2−4 HAU/50 μL | hemagglutin glycoprotein | DNA | 2017 | |
| [145] | H9N2 | Detection | RT-I-PCR | Kd = 40.67 nM | H9 gene | DNA | 26 nt | 2017 |
| [146] | H3N2 | Detection | Dual Recognition Element Lateral Flow Assay | 2 × 10−6 virus particles | A/Panama/2007/99 (H3N2) | RNA | 75 nt | 2014 |
| [122] | H5N2 | Therapy | 70% cell viability | hemagglutinin | RNA | 40 nt | 2014 | |
| [123] | H5N2 | Therapy | Selection and Binding | Significant inhibition of HA | hemagglutinin | RNA | 40 nt | 2011 |
| [125] | H9N2 | Therapy | capillary electrophoresis SELEX | H9N2 AIV purified haemagglutinin | DNA | - | 2015 | |
| [124] | H9N2 | Therapy | 3-fold survival | Same affinity as mouse antibody | globular region of the H9-type HA | DNA | 28 nt | 2011 |
| Reference | Virus | Virus Type | Detection/Therapy | Mode of Therapy/Detection | Detection Limit/Binding Affinity | Target | RNA/DNA | Size |
|---|---|---|---|---|---|---|---|---|
| [167] | Apple stem pitting virus | Detection | Molecularly Imprinted Polymer Gel Laser Diffraction Sensor | 10 ng/mL | MT32 protein | DNA | 40 nt | |
| [168] | bovine viral diarrhea virus | BVDV type 1 | Detection | Gold nano sandwich sensor | 800 copies/mL | Whole Virus | DNA | 30 nt |
| [169] | Coronavirus | SARS | Detection | Aptamer on Chip | 0.1 pg/mL | nucleocapsid protein | RNA | 92 nt |
| [170] | Norovirus | Human noroviruse | Detection | ELASA | NoV target-the P domain protein | DNA | ||
| [171] | Norovirus | GII.2 HuNoV strain, Snow Mountain Virus | Detection | Kd 191 nM | Immobilized SMV | DNA | 40 nt | |
| [172] | Norovirus | Murine Norovirus | Detection | Kd 290 nM G-quadruplexes | 20 aM to 120 aM | Viral SELEX | DNA | 40 nt |
| [173] | Porcine reproductive and respiratory syndrome virus | VR-2332 | Detection | Kd 2.5 × 105 TCID50/mL | In water 0.1 × 100–101 TCID50/mL Nasal/oral fluid 4.8 × 100–5.0 × 103 TCID50/mL | Viral SELEX | DNA | 40 nt |
| [174] | Vaccinia virus | Detection | Kd 26.3–40.9 nM | In water 60 PFU in 30 μL in blood 150 to 900 PFU | Viral SELEX | DNA | ||
| [175] | vaccinia virus | Detection | Impedimetric | 60 virions in a microliter | Virus Infected Cell SELEX | DNA | 40 nt | |
| [176] | vesicular stomatitis virus | Oncolytic Virus | Detection | To protect the Virus from Nuetralizing antibodies | 53–86% degree of Protection | Viral SELEX | DNA | 40 nt |
| [177] | VSV | vesicular stomatitis virus | Detection | aptamer-based affinity chromatography | switchable aptamers | |||
| [178] | Apple stem pitting virus isolates | Detection | ELONA | 81% affinity | MT32/PSA-H protein | DNA | 20-80 nt | |
| [179] | Bovine Herpes Virus-1 | Detection/Therapy | SELEX | 3.5 nM | gD protein | DNA | 38-51 nt | |
| [180] | Dengue Virus | Detection | SELEX | 5’-UTR | RNA | |||
| [181] | Human Papilloma virus (HPV) | Detection | SELEX | 10 nM | type 16 VLP | DNA | ||
| [182] | Iridovirus | Detection | ELISA | nmol L−1 | DNA | 71 nt | ||
| [183] | Parvovirus | Detection | SELEX | 467 nM | DNA | 49 nt | ||
| [184] | Respiratory Syncytial Virus (RSV) | Detection | SELEX | 0.5 nM | F protein | DNA | 30 nt | |
| [185] | Sindbis virus | Detection | ISH/RT-PCR | High affinity | E2 glycoprotein | RNA | ||
| [186] | Alphavirus | TC-83 Virus | Therapy | Replicon riobozyme actuator | Vaccine | RNA | ||
| [187] | Coronavirus | SARS | Therapy | Just binding studies | Kd 4–9 nM | nucleocapsid protein | DNA | 45 nt |
| [188] | Dengue | Therapy | DENV-2 envelop protein domain III (ED3) | DNA | 15 nt | |||
| [189] | Dengue | DENV-2 | Therapy | Thiolated aptamers | Kd 154 nM | envelope protein domain III | DNA | 24 nt |
| [190] | Ebola | VP35 | Therapy | Inhibition of VP35 | 10–50 nM | Viral Protein 35 | RNA | 45 nt |
| [191] | foot-and-mouth disease virus | Therapy | GFP tagged aptamers | 375 nM | RNA-dependent RNA polymerase | RNA | 30 nt | |
| [192] | Hemorrhagic Septicemia Virus | Therapy | Viral plaques were reduced by 50%, cells were viable and healthy | Viral SELEX | RNA | 40 nt | ||
| [193] | Hirame Rhabdovirus | Therapy | 15- to 63-fold reduction in plaques | Viral SELEX | RNA | 40 nt | ||
| [194] | HPV | HPV-16 | Therapy | Binding Inhibition | HPV-16 E7 protein | RNA | ||
| [195] | HPV | HPV-16 | Therapy | Kd = 0.05 pM | HPV-16 L1 VLPs | RNA | 15 nt | |
| [196] | HPV | HPV-16 | Therapy | Internalization of aptamers | 5 fold higher internalization | HPV-16 E6/E7 transformed tonsillar epithelial cell SELEX | RNA | 30 nt |
| [197] | Iridovirus | Singapore grouper | Therapy | inhibition | 12.09 nM | SGIV-infected GS cells | DNA | |
| [198] | Iridovirus | Singapore grouper | Therapy | 30% less mortality of fish | Whole Virus | DNA | 50 nt | |
| [199] | Norovirus | Therapy | capsid protein VP1 | DNA | 49 nt | |||
| [200] | rabies | Therapy | 25–33% survival | Kd 307 nM | RABV glycoprotein | DNA | 45 nt | |
| [201] | rabies | Therapy | 30–50% survival | Cell SELEX | DNA | |||
| [202] | rabies | Therapy | 4 nmol PEG-FO24 protected 87.5% | RABV-infected BHK-21 | DNA | 45 nt | ||
| [203] | rabies | Therapy | Kd 28–39 nM | Virus infected Cell SELEX | DNA | 45 nt | ||
| [204] | Rift Valley fever virus | Therapy | nucleocapsid protein | RNA | 30 nt | |||
| [205] | xenotropic murine leukemia virus-related virus (XMRV) | Different from HIV-1 | Therapy | IC50 2 nM | XMRV RT | RNA | three 12 nt sequences |
| Company | Custom Aptamers | SELEX Kits | Catalogue Aptamers | Random DNA Library | Clinical Trials | Country |
|---|---|---|---|---|---|---|
| IDT, 1710 Commercial Park, Coralville, Iowa 52241, USA | No | No | No | Yes | No | USA |
| AMSBio, 1035 Cambridge St Ste 16B, Cambridge, Massachusetts 02141-1154, USA | Yes | Yes | No | Yes | No | Spain |
| Gene Link, Inc. 8 Westchester Plaza, Suite 130, Elmsford, NY 10523, USA | Yes | No | No | Yes | No | USA |
| Roboklon, Robert-Rössle-Str. 10, 13125 Berlin, Germany | Yes | Yes | No | Yes | No | Germany |
| Ambiotech, 225 Broadway, #1903, New York, NY 10007, USA | Yes | Yes | No | Yes | No | USA |
| Jena Bioscience, Löbstedter Str. 71, 07749 Jena, Deutschland | Yes | No | No | Yes | No | Germany |
| TriLink, 10770 Wateridge Cir Ste 200, San Diego, CA 92121, USA | Yes | Yes | No | Yes | No | USA |
| IBA Lifesciences, Rudolf-Wissell-Str. 28, 37079 Göttingen, Germany | Yes | No | No | Yes | No | Germany |
| Neoventures, 516 Colborne St, London ON | Yes | No | No | Yes | No | Canada |
| OTC Biotech, San Antonio, Texas, USA | Yes | No | Yes | Yes | No | USA |
| Aptagen, Aptagen, LLC, 250 North Main Street, Jacobus, PA 17407, USA | Yes | No | Yes | Yes | No | USA |
| BasePair Technologies, 415 Madison Ave fl 4, New York, NY 10017, USA | Yes | No | Yes | Yes | No | USA |
| AptiSci, 301 Ho, 3 Dong, Pangyo Seven Venture Valley, Republic of Korea | Yes | No | Yes | Yes | No | South Korea |
| CamBio, 1 The Irwin Centre, Scotland Road, Dry Drayton, Cambridge, UK | No | No | From BasePair Technologies | Yes | No | UK |
| SomaLogic, 2945 Wilderness Pl. Boulder, CO 80301, USA | No | No | No | No | Yes | USA |
| Noxxon Pharma AG, Max-Dohrn-Strasse 8-10, 10589 Berlin, Germany | No | No | No | No | Yes | Germany |
| Gilead Sciences, Inc. 333 Lakeside Drive, Foster City, CA 94404, USA | No | No | No | No | Yes | USA |
| Sangon Biotech, 698 Xiangyu Road, Songjiang District, Shanghai Zip Code: 201611 | No | No | No | Yes | No | China |
| Takara Bio Inc, 2560 Orchard Parkway, San Jose, CA 95131, USA | No | No | No | Yes | No | China |
| AptaRes AG, Am Scheunenviertel 1, 15749 Mittenwalde, Germany | No | No | No | Yes | No | Germany |
| Bioneer, Inc. 155 Filbert St, Ste. 216, Oakland, CA 94607, USA | No | No | No | Yes | No | USA |
| Sigma Aldrich, St. Louis, MO 68178, USA | No | No | No | Yes | No | USA |
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Irvine, D.; Tuerk, C.; Gold, L. Selexion. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J. Mol. Biol. 1991, 222, 739–761. [Google Scholar] [CrossRef]
- Meng, H.-M.; Fu, T.; Zhang, X.-B.; Tan, W. Cell-SELEX-based aptamer-conjugated nanomaterials for cancer diagnosis and therapy. Natl. Sci. Rev. 2015, 2, 71–84. [Google Scholar] [CrossRef]
- Silha, J.V.; Murphy, L.J. The effects of the insulin-like growth factor-I aptamer, NBI-31772, on glucose homeostasis in the mouse. Can. J. Physiol. Pharmacol. 2005, 83, 557–563. [Google Scholar] [CrossRef]
- Nakamura, Y. Aptamer: Biology to applications. In Polydiacetylenes; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2011; pp. 135–152. [Google Scholar]
- Gedi, V.; Kim, Y.-P. Detection and characterization of cancer cells and pathogenic bacteria using aptamer-based nano-conjugates. Sensors 2014, 14, 18302–18327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Lü, Y.-H.; Yang, H.-Y.; Lin, J.-S.; Wang, Q.-Z. Recent progress of the aptamer-based antiviral drugs. Yao Xue Xue Bao Acta Pharm. Sin. 2013, 48, 447–456. [Google Scholar]
- Wilen, C.B.; Tilton, J.C.; Doms, R.W. HIV: Cell binding and entry. Cold Spring Harb. Perspect. Med. 2012, 2, a006866. [Google Scholar] [CrossRef]
- Chan, D.C.; Fass, D.; Berger, J.M.; Kim, P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997, 89, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Bugatti, A.; Giagulli, C.; Urbinati, C.; Caccuri, F.; Chiodelli, P.; Oreste, P.; Fiorentini, S.; Orro, A.; Milanesi, L.; D’Ursi, P.; et al. Molecular interaction studies of HIV-1 matrix protein p17 and heparin: Identification of the heparin-binding motif of p17 as a target for the development of multitarget antagonists. J. Biol. Chem. 2012, 288, 1150–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, A.L.; Donaghy, H.; Harman, A.N.; Kim, M.; Turville, S.G. Manipulation of dendritic cell function by viruses. Curr. Opin. Microbiol. 2010, 13, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Tang, M.; Zhang, M.; Majeed, S.; Montabana, E.; Stanfield, R.L.; Dimitrov, D.S.; Korber, B.; Sodroski, J.; Wilson, I.A.; et al. Structure of a V3-containing HIV-1 gp120 core. Science 2005, 310, 1025–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Lam, S.N.; Acharya, P.; Tang, M.; Xiang, S.; Hussan, S.S.; Stanfield, R.L.; Robinson, J.; Sodroski, J.; Wilson, I.A.; et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 2007, 317, 1930–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mushahwar, I.K. Human immunodeficiency viruses: Molecular virology, pathogenesis, diagnosis and treatment. Perspect. Med Virol. 2006, 13, 75–87. [Google Scholar] [CrossRef]
- Zhou, J.; Shu, Y.; Guo, P.; Smith, D.D.; Rossi, J.J. Dual functional RNA nanoparticles containing phi29 motor pRNA and anti-gp120 aptamer for cell-type specific delivery and HIV-1 inhibition. Methods 2011, 54, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Neff, C.P.; Swiderski, P.; Li, H.; Smith, D.D.; Aboellail, T.; Remling-Mulder, L.; Akkina, R.; Rossi, J.J. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 2013, 21, 192–200. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Rossi, J.J. Aptamer-targeted RNAi for HIV-1 therapy. Methods Mol. Biol. 2011, 721, 355–371. [Google Scholar] [CrossRef]
- Wheeler, L.A.; Trifonova, R.; Vrbanac, V.; Basar, E.; McKernan, S.; Xu, Z.; Seung, E.; Deruaz, M.; Dudek, T.; Einarsson, J.I.; et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Investig. 2011, 121, 2401–2412. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, L.A.; Vrbanac, V.; Trifonova, R.; Brehm, M.A.; Gilboa-Geffen, A.; Tanno, S.; Greiner, D.L.; Luster, A.D.; Tager, A.M.; Lieberman, J. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol. Ther. 2013, 21, 1378–1389. [Google Scholar] [CrossRef] [Green Version]
- Mufhandu, H.T.; Gray, E.S.; Madiga, M.C.; Tumba, N.; Alexandre, K.B.; Khoza, T.; Wibmer, C.K.; Moore, P.L.; Morris, L.; Khati, M. UCLA1, a synthetic derivative of a gp120 RNA aptamer, inhibits entry of human immunodeficiency virus type 1 subtype C. J. Virol. 2012, 86, 4989–4999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes de Campos, W.R.; Chirwa, N.; London, G.; Rotherham, L.S.; Morris, L.; Mayosi, B.M.; Khati, M. HIV-1 subtype C unproductively infects human cardiomyocytes in vitro and induces apoptosis mitigated by an anti-gp120 aptamer. PLoS ONE 2014, 9, e110930. [Google Scholar] [CrossRef] [Green Version]
- London, G.M.; Mayosi, B.M.; Khati, M. Isolation and characterization of 2′-F-RNA aptamers against whole HIV-1 subtype C envelope pseudovirus. Biochem. Biophys. Res. Commun. 2015, 456, 428–433. [Google Scholar] [CrossRef]
- Musumeci, D.; Riccardi, C.; Montesarchio, D. G-quadruplex forming oligonucleotides as anti-HIV agents. Molecules 2015, 20, 17511–17532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyatt, J.R.; Vickers, T.A.; Roberson, J.L.; Buckheit, R.W.; Klimkait, T.; DeBaets, E.; Davis, P.W.; Rayner, B.; Imbach, J.L.; Ecker, D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. USA 1994, 91, 1356–1360. [Google Scholar] [CrossRef] [Green Version]
- Koizumi, M.; Koga, R.; Hotoda, H.; Momota, K.; Ohmine, T.; Furukawa, H.; Agatsuma, T.; Nishigaki, T.; Abe, K.; Kosaka, T.; et al. Biologically active oligodeoxyribonucleotides—IX.1 synthesis and anti-HIV-1 activity of hexadeoxyribonucleotides, TGGGAG, bearing 3′- and 5′-end-modification. Bioorganic Med. Chem. 1997, 5, 2235–2243. [Google Scholar] [CrossRef]
- Hotoda, H.; Koizumi, M.; Koga, R.; Kaneko, M.; Momota, K.; Ohmine, T.; Furukawa, H.; Agatsuma, T.; Nishigaki, T.; Sone, J.; et al. Biologically active oligodeoxyribonucleotides. 5. 5′-end-substituted d(TGGGAG) possesses anti-human immunodeficiency virus type 1 activity by forming a G-quadruplex structure. J. Med. Chem. 1998, 41, 3655–3663. [Google Scholar] [CrossRef]
- D’Onofrio, J.; Petraccone, L.; Erra, E.; Martino, L.; Di Fabio, G.; de Napoli, L.; Giancola, C.; Montesarchio, D. 5′-modified G-quadruplex forming oligonucleotides endowed with anti-HIV activity: Synthesis and biophysical properties. Bioconjugate Chem. 2007, 18, 1194–1204. [Google Scholar] [CrossRef]
- D’Onofrio, J.; Petraccone, L.; Martino, L.; Di Fabio, G.; Iadonisi, A.; Balzarini, J.; Giancola, C.; Montesarchio, D. Synthesis, biophysical characterization, and anti-HIV activity of glyco-conjugated G-quadruplex-forming oligonucleotides. Bioconjugate Chem. 2008, 19, 607–616. [Google Scholar] [CrossRef]
- Di Fabio, G.; D’Onofrio, J.; Chiapparelli, M.; Hoorelbeke, B.; Montesarchio, D.; Balzarini, J.; Napoli, L.D. Discovery of novel anti-HIV active G-quadruplex-forming oligonucleotides. Chem. Commun. 2011, 47, 2363–2365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanucci, V.; Gaglione, M.; Messere, A.; Potenza, N.; Zarrelli, A.; Noppen, S.; Liekens, S.; Balzarini, J.; Di Fabio, G. Hairpin oligonucleotides forming G-quadruplexes: New aptamers with anti-HIV activity. Eur. J. Med. Chem. 2014, 89, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Romanucci, V.; Milardi, D.; Campagna, T.; Gaglione, M.; Messere, A.; D’Urso, A.; Crisafi, E.; la Rosa, C.; Zarrelli, A.; Balzarini, J.; et al. Synthesis, biophysical characterization and anti-HIV activity of d(TG 3AG) quadruplexes bearing hydrophobic tails at the 5′-end. Bioorg. Med. Chem. 2014, 22, 960–966. [Google Scholar] [CrossRef]
- D’Atri, V.; Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Mayol, L.; Piccialli, V.; Haider, S.; Hoorelbeke, B.; Balzarini, J.; et al. New anti-HIV aptamers based on tetra-end-linked DNA G-quadruplexes: Effect of the base sequence on anti-HIV activity. Chem. Commun. 2012, 48, 9516–9518. [Google Scholar] [CrossRef]
- Nici, F.; Oliviero, G.; Falanga, A.P.; D’Errico, S.; Marzano, M.; Musumeci, D.; Montesarchio, D.; Noppen, S.; Pannecouque, C.; Piccialli, G. Anti-HIV activity of new higher order G-quadruplex aptamers obtained from tetra-end-linked oligonucleotides. Org. Biomol. Chem. 2018, 16, 2349–2355. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, A.; Esposito, V.; Citarella, G.; Mayol, L.; Galeone, A. Structural investigations on the anti-HIV G-quadruplex-forming oligonucleotide TGGGAG and its analogues: Evidence for the presence of an A-tetrad. ChemBioChem 2012, 13, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.J.; Sharma, T.K.; Whatley, A.S.; Landon, L.A.; Tempesta, M.A.; Johnson, M.C.; Burke, D.H. Robust suppression of HIV replication by intracellularly expressed reverse transcriptase aptamers is independent of ribozyme processing. Mol. Ther. 2012, 20, 2304–2314. [Google Scholar] [CrossRef] [Green Version]
- Ditzler, M.A.; Bose, D.; Shkriabai, N.; Marchand, B.; Sarafianos, S.G.; Kvaratskhelia, M.; Burke, D.H. Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates. Nucleic Acids Res. 2011, 39, 8237–8247. [Google Scholar] [CrossRef] [PubMed]
- Whatley, A.S.; Ditzler, M.A.; Lange, M.J.; Biondi, E.; Sawyer, A.W.; Chang, J.L.; Franken, J.D.; Burke, D.H. Potent inhibition of HIV-1 reverse transcriptase and replication by nonpseudoknot, “UCAA-motif” RNA aptamers. Mol. Ther. Nucl. Acids 2013, 2, e71. [Google Scholar] [CrossRef]
- Lange, M.J.; Burke, D.H. Screening inhibitory potential of anti-HIV RT RNA aptamers. Methods Mol. Biol. 2014, 1103, 11–29. [Google Scholar]
- Ruslinda, A.R.; Wang, X.; Ishii, Y.; Ishiyama, Y.; Tanabe, K.; Kawarada, H. Human immunodeficiency virus trans-activator of transcription peptide detection via ribonucleic acid aptamer on aminated diamond biosensor. Appl. Phys. Lett. 2011, 99, 123702. [Google Scholar] [CrossRef]
- Ruslinda, A.R.; Tanabe, K.; Ibori, S.; Wang, X.; Kawarada, H. Effects of diamond-FET-based RNA aptamer sensing for detection of real sample of HIV-1 tat protein. Biosens Bioelectron. 2013, 40, 277–282. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, Z.; Wei, H.; Hu, Q.; Deng, J.; Guo, D.; Cui, Z.; Zhang, X.-E. Aptamer beacons for visualization of endogenous protein HIV-1 reverse transcriptase in living cells. Biosens. Bioelectron. 2011, 28, 270–276. [Google Scholar] [CrossRef]
- Kim, M.-G.; Shon, Y.; Lee, J.; Byun, Y.; Choi, B.-S.; Kim, Y.B.; Oh, Y.-K. Double stranded aptamer-anchored reduced graphene oxide as target-specific nano detector. Biomaterials 2014, 35, 2999–3004. [Google Scholar] [CrossRef]
- Niedzwiecki, D.J.; Iyer, R.; Borer, P.N.; Movileanu, L. Sampling a biomarker of the human immunodeficiency virus across a synthetic nanopore. ACS Nano 2013, 7, 3341–3350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, Y.D.; Bukharina, N.S.; Pleshakova, T.O.; Frantsuzov, P.A.; Andreeva, E.Y.; Kaysheva, A.L.; Zgoda, V.G.; Izotov, A.A.; Pavlova, T.I.; Ziborov, V.S.; et al. Atomic force microscopy fishing and mass spectrometry identification of gp120 on immobilized aptamers. Int. J. Nanomed. 2014, 9, 4659–4670. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Bai, X.; Wen, W.; Zhang, X.; Wang, S. Ultrasensitive electrochemical biosensor for HIV gene detection based on graphene stabilized gold nanoclusters with exonuclease amplification. ACS Appl. Mater. Interfaces 2015, 7, 18872–18879. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Bravo, I.A.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar] [CrossRef] [Green Version]
- Esposito, V.; Pirone, L.; Mayol, L.; Pedone, E.; Virgilio, A.; Galeone, A. Exploring the binding of d(GGGT)4 to the HIV-1 integrase: An approach to investigate G-quadruplex aptamer/target protein interactions. Biochimie 2016, 127, 19–22. [Google Scholar] [CrossRef] [Green Version]
- Mufhandu, H.T.; Alexandre, K.B.; Gray, E.S.; Morris, L.; Khati, M. UCLA1 aptamer inhibition of human immunodeficiency virus type 1 subtype C primary isolates in macrophages and selection of resistance. Biochem. Biophys. Rep. 2016, 7, 408–414. [Google Scholar] [CrossRef] [Green Version]
- Lange, M.J.; Nguyen, P.D.M.; Callaway, M.K.; Johnson, M.C.; Burke, D.H. RNA-protein interactions govern antiviral specificity and encapsidation of broad spectrum anti-HIV reverse transcriptase aptamers. Nucleic Acids Res. 2017, 45, 6087–6097. [Google Scholar] [CrossRef] [Green Version]
- Prokofjeva, M.; Tsvetkov, V.; Basmanov, D.; Varizhuk, A.; Lagarkova, M.; Smirnov, I.; Prusakov, K.; Klinov, D.; Prassolov, V.; Pozmogova, G.; et al. Anti-HIV activities of intramolecular G4 and non-G4 oligonucleotides. Nucleic Acid Ther. 2017, 27, 56–66. [Google Scholar] [CrossRef]
- Yamaoki, Y.; Nagata, T.; Mashima, T.; Katahira, M. Development of an RNA aptamer that acquires binding capacity against HIV-1 tat protein: Via G-quadruplex formation in response to potassium ions. Chem. Commun. 2017, 53, 7056–7059. [Google Scholar] [CrossRef]
- Métifiot, M.; Amrane, S.; Mergny, J.-L.; Andreola, M.-L. Anticancer molecule AS1411 exhibits low nanomolar antiviral activity against HIV-1. Biochimie 2015, 118, 173–175. [Google Scholar] [CrossRef]
- Catuogno, S.; Esposito, C.L.; de Franciscis, V. A trojan horse for human immunodeficiency virus. Chem. Biol. 2015, 22, 313–314. [Google Scholar] [CrossRef] [Green Version]
- Duclair, S.; Gautam, A.; Ellington, A.; Prasad, V.R. High-affinity RNA aptamers against the HIV-1 protease inhibit both in vitro protease activity and late events of viral replication. Mol. Ther. Nucl. Acids 2015, 4, e228. [Google Scholar] [CrossRef]
- Takahashi, M.; Burnett, J.C.; Rossi, J.J. Aptamer–siRNA chimeras for HIV. Adv. Exp. Med. Biol. 2015, 848, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Luque, F.J.; Stich, M.; Manrubia, S.; Briones, C.; Berzal-Herranz, A. Efficient HIV-1 inhibition by a 16 nt-long RNA aptamer designed by combining in vitro selection and in silico optimisation strategies. Sci. Rep. 2014, 4, srep06242. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Pei, S.-N.; Parekh, P.; Salazar, E.; Zu, Y. Blocking interaction of viral gp120 and CD4-expressing T cells by single-stranded DNA aptamers. Int. J. Biochem. Cell Biol. 2014, 51, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Shiang, Y.-C.; Ou, C.-M.; Chen, S.-J.; Ou, T.-Y.; Lin, H.-J.; Huang, C.-C.; Chang, H.-T. Highly efficient inhibition of human immunodeficiency virus type 1 reverse transcriptase by aptamers functionalized gold nanoparticles. Nanoscale 2013, 5, 2756–2764. [Google Scholar] [CrossRef]
- Xu, L.; Cai, L.; Chen, X.; Jiang, X.; Chong, H.; Zheng, B.; Wang, K.; He, J.; Chen, W.; Zhang, T.; et al. DNA duplexes with hydrophobic modifications inhibit fusion between HIV-1 and cell membranes. Antimicrob. Agents Chemother. 2013, 57, 4963–4970. [Google Scholar] [CrossRef] [Green Version]
- Zhan, S.B.; Zhang, X.G.; Li, H.X.; Zeng, Y. A novel SELEX method for screening of HIV-1 P24 antigen. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2013, 27, 218–220. [Google Scholar]
- Possik, E.J.; Bou Sleiman, M.S.; Ghattas, I.R.; Smith, C.A. Randomized codon mutagenesis reveals that the HIV rev arginine-rich motif is robust to substitutions and that double substitution of two critical residues alters specificity. J. Mol. Recognit. 2013, 26, 286–296. [Google Scholar] [CrossRef]
- Magbanua, E.; Zivkovic, T.; Hansen, B.; Beschorner, N.; Meyer, C.; Lorenzen, I.; Grötzinger, J.; Hauber, J.; Torda, A.E.; Mayer, G.; et al. D(GGGT)4 and r(GGGU)4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biology 2013, 10, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Shibata, T.; Kabashima, T.; Kai, M. Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA. Eur. J. Med. Chem. 2012, 56, 396–399. [Google Scholar] [CrossRef]
- Scheideman, E.H.; Marlatt, S.A.; Xie, Y.; Hu, Y.; Sutton, R.E.; Dimaio, D. Transmembrane protein aptamers that inhibit CCR5 expression and HIV coreceptor function. J. Virol. 2012, 86, 10281–10292. [Google Scholar] [CrossRef] [Green Version]
- Um, H.-J.; Kim, M.; Lee, S.-H.; Kim, Y.-H. Preventing the formation of positive transcription elongation factor b by human cyclin T1-binding RNA aptamer for anti-HIV transcription. AIDS 2012, 26, 1599–1605. [Google Scholar] [CrossRef]
- Lai, Y.-T.; DeStefano, J.J. A primer-free method that selects high-affinity single-stranded DNA aptamers using thermostable RNA ligase. Anal. Biochem. 2011, 414, 246–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faure-Perraud, A.; Métifiot, M.; Reigadas, S.; Recordon-Pinson, P.; Parissi, V.; Ventura, M.; Andréola, M.-L. The guanine-quadruplex aptamer 93del inhibits HIV-1 replication ex vivo by interfering with viral entry, reverse transcription and integration. Antivir. Ther. 2011, 16, 383–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Satheesan, S.; Li, H.; Weinberg, M.S.; Morris, K.V.; Burnett, J.C.; Rossi, J.J. Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem. Biol. 2015, 22, 379–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neff, C.P.; Zhou, J.; Remling, L.; Kuruvilla, J.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Rossi, J.J.; Akkina, R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4+ T cell decline in humanized mice. Sci. Transl. Med. 2011, 3, 66ra6. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, D.; Duclair, S.; Datta, S.A.K.; Ellington, A.; Rein, A.; Prasad, V.R. RNA aptamers directed to human immunodeficiency virus type 1 gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011, 85, 305–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watashi, K.; Urban, S.; Li, W.; Wakita, T. NTCP and beyond: Opening the door to unveil hepatitis B virus entry. Int. J. Mol. Sci. 2014, 15, 2892–2905. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization (WHO). Hepatits. Available online: http://www.who.int/csr/disease/hepatitis/whocdscsrlyo20022/en/index8.html#23 (accessed on 20 April 2015).
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012, 1, e00049. [Google Scholar] [CrossRef]
- Gao, W.; Hu, J. Formation of hepatitis B virus covalently closed circular DNA: Removal of genome-linked protein. J. Virol. 2007, 81, 6164–6174. [Google Scholar] [CrossRef] [Green Version]
- Lindenbach, B.D.; Rice, C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013, 11, 688–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- André, P.; Pradel, F.; Deforges, S.; Perret, M.; Berland, J.-L.; Sodoyer, M.; Pol, S.; Bréchot, C.; Paranhos-Baccalà, G.; Lotteau, V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 2002, 76, 7040–7048. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, E.; Belouzard, S.; Goueslain, L.; Wakita, T.; Dubuisson, J.; Wychowski, C.; Rouillé, Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 2006, 80, 6964–6972. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Qian, X.; Zhao, P.; Qi, Z. How hepatitis C virus invades hepatocytes: The mystery of viral entry. World J. Gastroenterol. 2014, 20, 3457–3467. [Google Scholar] [CrossRef] [PubMed]
- Coller, K.E.; Heaton, N.S.; Berger, K.L.; Cooper, J.D.; Saunders, J.L.; Randall, G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012, 8, e1002466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Beck, J.; Nassal, M.; Hu, K.-H. A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS ONE 2011, 6, e27862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; T-Thienprasert, N.P.; Brown, C.M. Prospects for inhibiting the post-transcriptional regulation of gene expression in hepatitis B virus. World J. Gastroenterol. 2014, 20, 7993–8004. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Pei, X.; Zhang, Q.; Lu, B.; Zhang, X.; Liu, J. An aptamer targets HBV core protein and suppresses HBV replication in HepG2.2.15 cells. Int. J. Mol. Med. 2014, 34, 1423–1429. [Google Scholar] [CrossRef] [Green Version]
- Orabi, A.; Bieringer, M.; Geerlof, A.; Bruss, V. An aptamer against the matrix binding domain on the hepatitis B virus capsid impairs virion formation. J. Virol. 2015, 89, 9281–9287. [Google Scholar] [CrossRef] [Green Version]
- Marton, S.; Berzal-Herranz, B.; Garmendia, E.; Cueto, F.J.; Berzal-Herranz, A. Anti-HCV RNA aptamers targeting the genomic cis-acting replication element. Pharmaceuticals 2011, 5, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-López, C.; Berzal-Herranz, B.; Gómez, J.; Berzal-Herranz, A. An engineered inhibitor RNA that efficiently interferes with hepatitis C virus translation and replication. Antivir. Res. 2012, 94, 131–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marton, S.; Romero-Lõpez, C.; Berzal-Herranz, A. RNA aptamer-mediated interference of HCV replication by targeting the CRE-5BSL3.2 domain. J. Viral Hepat. 2013, 20, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.H.; Lee, S.-W. Inhibition of hepatitis C virus (HCV) replication by hammerhead ribozyme which activity can be allosterically regulated by HCV NS5B RNA replicase. Korean J. Microbiol. 2011, 47, 188–193. [Google Scholar]
- Lee, C.H.; Kim, J.H.; Lee, S.-W. Prospects for nucleic acid-based therapeutics against hepatitis C virus. World J. Gastroenterol. 2013, 19, 8949–8962. [Google Scholar] [CrossRef]
- Lee, C.H.; Lee, S.-H.; Kim, J.H.; Noh, Y.-H.; Noh, G.-J.; Lee, S.-W. Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis C virus (HCV) NS5B protein. Mol. Ther. Nucl. Acids 2015, 4, e254. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Lee, Y.J.; Kim, J.H.; Lim, J.H.; Kim, J.-H.; Han, W.; Lee, S.-H.; Noh, G.-J.; Lee, S.-W. Inhibition of hepatitis C virus (HCV) replication by specific RNA aptamers against HCV NS5B RNA replicase. J. Virol. 2013, 87, 7064–7074. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Meng, X.; Yu, Q.; Xu, L.; Long, Y.; Liu, B.; Fang, X.; Zhu, H. Inhibition of hepatitis C virus infection by DNA aptamer against envelope protein. Antimicrob. Agents Chemother. 2013, 57, 4937–4944. [Google Scholar] [CrossRef] [Green Version]
- Delaviz, N.; Gill, P.; Ajami, A.; Aarabi, M. Aptamer-conjugated magnetic nanoparticles for the efficient removal of HCV particles from human plasma samples. RSC Adv. 2015, 5, 79433–79439. [Google Scholar] [CrossRef]
- Suh, S.-K.; Song, S.; Oh, H.-B.; Hwang, S.; Hah, S.S. Aptamer-based competitive binding assay for one-step quantitation of hepatitis B surface antigen. Analyst 2014, 139, 4310–4314. [Google Scholar] [CrossRef]
- Xi, Z.; Huang, R.; Li, Z.; He, N.; Wang, T.; Su, E.; Deng, Y. Selection of HBsAg-specific DNA aptamers based on carboxylated magnetic nanoparticles and their application in the rapid and simple detection of hepatitis b virus infection. ACS Appl. Mater. Interfaces 2015, 7, 11215–11223. [Google Scholar] [CrossRef]
- Roh, C.; Kim, S.E.; Jo, S.-K. Label free inhibitor screening of hepatitis C virus (HCV) NS5B viral protein using RNA oligonucleotide. Sensors 2011, 11, 6685–6696. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, L.; Shen, X. Development of a nucleic acid lateral flow strip for detection of hepatitis C virus (HCV) core antigen. Nucleosides Nucleotides Nucleic Acids 2013, 32, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jee, M.H.; Kwon, O.S.; Keum, S.J.; Jang, S.K. Infectivity of hepatitis C virus correlates with the amount of envelope protein E2: Development of a new aptamer-based assay system suitable for measuring the infectious titer of HCV. Virology 2013, 439, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Chen, S.-C.; Zhou, J.; Chen, Z.-D.; Chen, F. Identification of aptamer-binding sites in hepatitis C virus envelope glycoprotein E2. Iran J. Med. Sci. 2015, 40, 63–67. [Google Scholar] [PubMed]
- Bang, G.S.; Cho, S.; Lee, N.; Lee, B.-R.; Kim, J.-H.; Kim, B.-G. Rational design of modular allosteric aptamer sensor for label-free protein detection. Biosens. Bioelectron. 2013, 39, 44–50. [Google Scholar] [CrossRef]
- Huang, R.; Xi, Z.; Deng, Y.; He, N. Fluorescence based Aptasensors for the determination of hepatitis B virus e antigen. Sci. Rep. 2016, 6, 31103. [Google Scholar] [CrossRef] [Green Version]
- Qu, F.; Liu, Y.; Kong, R.; You, J. A versatile DNA detection scheme based on the quenching of fluorescent silver nanoclusters by MoS2nanosheets: Application to aptamer-based determination of hepatitis B virus and of dopamine. Microchim. Acta 2017, 184, 4417–4424. [Google Scholar] [CrossRef]
- Liu, G.; Li, J.; Feng, D.-Q.; Zhu, J.-J.; Wang, W. Silver nanoclusters beacon as stimuli-responsive versatile platform for multiplex DNAs detection and aptamer-substrate complexes sensing. Anal. Chem. 2017, 89, 1002–1008. [Google Scholar] [CrossRef]
- Ghanbari, K.; Roushani, M.; Azadbakht, A. Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 2017, 534, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Kaysheva, A.L.; Bayzyanova, J.M.; Anashkina, A.S.; Uchaikin, V.F.; Ziborov, V.S.; Konev, V.A.; Archakov, A.I.; Ivanov, Y.D. The detection of hepatitis C virus core antigen using afm chips with immobolized aptamers. J. Virol. Methods 2018, 251, 99–105. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Cheng, H.; Sun, N.; Liu, M.; Wu, Z.; Pei, R. Inducible Bcl-2 gene RNA interference mediated by aptamer-integrated HDV ribozyme switch. Integr. Biol. 2017, 9, 619–626. [Google Scholar] [CrossRef]
- Nomura, Y.; Zhou, L.; Miu, A.; Yokobayashi, Y. Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth. Biol. 2013, 2, 684–689. [Google Scholar] [CrossRef]
- Gao, Y.; Yu, X.; Xue, B.; Zhou, F.; Wang, X.; Yang, D.; Liu, N.; Xu, L.; Fang, X.; Zhu, H. Inhibition of hepatitis C virus infection by dna aptamer against NS2 protein. PLoS ONE 2014, 9, e90333. [Google Scholar] [CrossRef]
- Shi, S.; Yu, X.; Gao, Y.; Xue, B.; Wu, X.; Wang, X.; Yang, D.; Zhu, H. Inhibition of hepatitis C virus production by aptamers against the core protein. J. Virol. 2014, 88, 1990–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Gao, Y.; Xue, B.; Wang, X.; Yang, D.; Qin, Y.; Yu, R.; Liu, N.; Xu, L.; Fang, X.; et al. Inhibition of hepatitis C virus infection by NS5A-specific aptamer. Antivir. Res. 2014, 106, 116–124. [Google Scholar] [CrossRef]
- Rapid Reference Influenza. Available online: http://www.rapidreferenceinfluenza.com/resource-center (accessed on 20 April 2016).
- Samji, T. Influenza A: Understanding the viral life cycle. Yale J. Biol. Med. 2009, 82, 153–159. [Google Scholar] [PubMed]
- Matrosovich, M.; Tuzikov, A.; Bovin, N.; Gambaryan, A.; Klimov, A.; Castrucci, M.R.; Donatelli, I.; Kawaoka, Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 2000, 74, 8502–8512. [Google Scholar] [CrossRef] [Green Version]
- Lakadamyali, M.; Rust, M.J.; Zhuang, X. Endocytosis of influenza viruses. Microbes Infect 2004, 6, 929–936. [Google Scholar] [CrossRef] [Green Version]
- Shtyrya, Y.A.; Mochalova, L.V.; Bovin, N.V. Influenza virus neuraminidase: Structure and function. Acta Nat. 2009, 1, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, S.C.B.; Kumar, P.K.R. Aptamers that bind to the hemagglutinin of the recent pandemic influenza virus H1N1 and efficiently inhibit agglutination. Acta Biomater. 2013, 9, 8932–8941. [Google Scholar] [CrossRef] [PubMed]
- Musafia, B.; Oren-Banaroya, R.; Noiman, S. Designing anti-influenza aptamers: Novel quantitative structure activity relationship approach gives insights into aptamer—Virus interaction. PLoS ONE 2014, 9, e97696. [Google Scholar] [CrossRef] [PubMed]
- Wongphatcharachai, M.; Wang, P.; Enomoto, S.; Webby, R.J.; Gramer, M.R.; Amonsin, A.; Sreevatsan, S. Neutralizing DNA aptamers against swine influenza H3N2 viruses. J. Clin. Microbiol. 2013, 51, 46–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, H.-M.; Lee, J.-M.; Yim, S.; Jeong, Y.-J. Isolation of single-stranded DNA aptamers that distinguish influenza virus hemagglutinin subtype H1 from H5. PLoS ONE 2015, 10, e0125060. [Google Scholar] [CrossRef] [Green Version]
- Suenaga, E.; Kumar, P.K.R. An aptamer that binds efficiently to the hemagglutinins of highly pathogenic avian influenza viruses (H5N1 and H7N7) and inhibits hemagglutinin-glycan interactions. Acta Biomater. 2014, 10, 1314–1323. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Zhang, N.; Singh, K.; Shuai, H.; Chu, H.; Zhou, J.; Chow, B.K.C.; Zheng, B.-J. Cross-protection of influenza A virus infection by a DNA aptamer targeting the PA endonuclease domain. Antimicrob. Agents Chemother. 2015, 59, 4082–4093. [Google Scholar] [CrossRef] [Green Version]
- Kwon, H.-M.; Lee, K.H.; Han, B.W.; Han, M.R.; Kim, D.H. An RNA aptamer that specifically binds to the glycosylated hemagglutinin of avian influenza virus and suppresses viral infection in cells. PLoS ONE 2014, 9, e97574. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Kim, S.; Yoon, H.; Kim, K.-B.; Kalme, S.S.; Oh, S.; Song, C.S.; Kim, D.-E. Selection of an antiviral RNA aptamer against hemagglutinin of the subtype H5 avian influenza virus. Nucleic Acid Ther. 2011, 21, 395–402. [Google Scholar] [CrossRef]
- Choi, S.K.; Lee, C.; Lee, K.S.; Choe, S.-Y.; Mo, I.P.; Seong, R.H.; Hong, S.; Jeon, S.H. DNA aptamers against the receptor binding region of hemagglutinin prevent avian influenza viral infection. Mol. Cells 2011, 32, 527–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yu, Z.; Jiang, F.; Fu, P.; Shen, J.; Wu, W.; Li, J. Two DNA aptamers against avian influenza H9N2 virus prevent viral infection in cells. PLoS ONE 2015, 10, e0123060. [Google Scholar] [CrossRef]
- Bai, H.; Wang, R.; Hargis, B.; Lu, H.; Li, Y. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors 2012, 12, 12506–12518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Li, Y. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 2013, 42, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, J.; Jiang, T.; Kwon, Y.M.; Lu, H.; Jiao, P.; Liao, M.; Li, Y. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J. Virol. Methods 2013, 189, 362–369. [Google Scholar] [CrossRef]
- Fu, Y.; Callaway, Z.; Lum, J.; Wang, R.; Lin, J.; Li, Y. Exploiting enzyme catalysis in ultra-low ion strength media for impedance biosensing of avian influenza virus using a bare interdigitated electrode. Anal. Chem. 2014, 86, 1965–1971. [Google Scholar] [CrossRef]
- Wang, R.; Xu, L.; Li, Y. Bio-nanogate controlled enzymatic reaction for virus sensing. Biosens. Bioelectron. 2015, 67, 400–407. [Google Scholar] [CrossRef]
- Lum, J.; Wang, R.; Hargis, B.; Tung, S.; Bottje, W.; Lu, H.; Li, Y. An impedance aptasensor with microfluidic chips for specific detection of H5N1 avian influenza virus. Sensors 2015, 15, 18565–18578. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Cheng, Z.; Fan, H.; Ai, S.; Han, R. Electrochemical detection of avian influenza virus H5N1 gene sequence using a DNA aptamer immobilized onto a hybrid nanomaterial-modified electrode. Electrochim. Acta 2011, 56, 6266–6270. [Google Scholar] [CrossRef]
- Diba, F.S.; Kim, S.; Lee, H.J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015, 72, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Hushegyi, A.; Pihíková, D.; Bertok, T.; Adam, V.; Kizek, R.; Tkac, J. Ultrasensitive detection of influenza viruses with a glycan-based impedimetric biosensor. Biosens. Bioelectron. 2016, 79, 644–649. [Google Scholar] [CrossRef] [Green Version]
- Negri, P.; Chen, G.; Kage, A.; Nitsche, A.; Naumann, D.; Xu, B.; Dluhy, R.A. Direct optical detection of viral nucleoprotein binding to an anti-influenza aptamer. Anal. Chem. 2012, 84, 5501–5508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negri, P.; Kage, A.; Nitsche, A.; Naumann, D.; Dluhy, R.A. Detection of viral nucleoprotein binding to anti-influenza aptamers via SERS. Chem. Commun. 2011, 47, 8635–8637. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.-C.; Wang, C.-H.; Liou, T.-M.; Lee, G.-B. Influenza A virus-specific aptamers screened by using an integrated microfluidic system. Lab Chip 2014, 14, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Adamiak, B.; Benton, D.J.; Johnson, C.J.; Sharma, S.; Fenton, R.; McCauley, J.W.; Iqbal, M.; Cass, A.E.G. Aptamer-based biosensors for the rapid visual detection of flu viruses. Chem. Commun. 2014, 50, 15533–15536. [Google Scholar] [CrossRef] [PubMed]
- Lakshmipriya, T.; Fujimaki, M.; Gopinath, S.C.B.; Awazu, K. Generation of anti-influenza aptamers using the systematic evolution of ligands by exponential enrichment for sensing applications. Langmuir 2013, 29, 15107–15115. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Rong, Z.; Wang, J.; Xiao, R.; Wang, S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF). Biosens. Bioelectron. 2015, 66, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, I.; Akitomi, J.; Boltz, D.A.; Horii, K.; Furuichi, M.; Waga, I. Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014, 443, 37–41. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.Q.; Ren, Q.; Wei, H.P.; Chen, Z.; Deng, J.Y.; Zhang, Z.P.; Zhang, X.E. Quantum dot-aptamer nanoprobes for recognizing and labeling influenza A virus particles. Nanoscale 2011, 3, 2454–2457. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-T.; Wang, C.-H.; Chang, C.-P.; Lee, G.-B. Integrated microfluidic system for rapid detection of influenza H1N1 virus using a sandwich-based aptamer assay. Biosens. Bioelectron. 2016, 82, 105–111. [Google Scholar] [CrossRef]
- Wang, R.; Wang, L.; Callaway, Z.T.; Lu, H.; Huang, T.J.; Li, Y. A nanowell-based QCM aptasensor for rapid and sensitive detection of avian influenza virus. Sens. Actuators B Chem. 2017, 240, 934–940. [Google Scholar] [CrossRef] [Green Version]
- Hmila, I.; Wongphatcharachai, M.; Laamiri, N.; Aouini, R.; Marnissi, B.; Arbi, M.; Sreevatsan, S.; Ghram, A. A novel method for detection of H9N2 influenza viruses by an aptamer-real time-PCR. J. Virol. Methods 2017, 243, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Chang, P.; Benton, D.J.; McCauley, J.W.; Iqbal, M.; Cass, A.E.G. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017, 89, 6781–6786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilling, A.; Rosenberg, M.F.; Willis, S.H.; Jäger, J.; Cohen, G.H.; Eisenberg, R.J.; Meredith, D.M.; Holzenburg, A. Three-dimensional structure of herpes simplex virus type 1 glycoprotein D at 2.4-nanometer resolution. J. Virol. 1999, 73, 7830–7834. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, J.; Shukla, D. Viral entry mechanisms: Cellular and viral mediators of herpes simplex virus entry. FEBS J 2009, 276, 7228–7236. [Google Scholar] [CrossRef]
- Eisenberg, R.J.; Atanasiu, D.; Cairns, T.M.; Gallagher, J.R.; Krummenacher, C.; Cohen, G.H. Herpes virus fusion and entry: A story with many characters. Viruses 2012, 4, 800–832. [Google Scholar] [CrossRef]
- Haarr, L.; Skulstad, S. The herpes simplex virus type 1 particle: Structure and molecular functions. APMIS 1994, 102, 321–346. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, W.; Huang, C.C.; Herr, W.; Cheng, X. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 1999, 13, 1692–1703. [Google Scholar] [CrossRef] [Green Version]
- Li, M.-L.; Guo, H.; Ding, Q.; Zheng, C.-F. A multiple functional protein: The herpes simplex virus type 1 tegument protein VP22. Virol. Sin. 2009, 24, 153–161. [Google Scholar] [CrossRef]
- Long, D.; Wilcox, W.C.; Abrams, W.R.; Cohen, G.H.; Eisenberg, R.J. Disulfide bond structure of glycoprotein D of herpes simplex virus types 1 and 2. J. Virol. 1992, 66, 6668–6685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heldwein, E.E.; Lou, H.; Bender, F.C.; Cohen, G.H.; Eisenberg, R.J.; Harrison, S.C. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 2006, 313, 217–220. [Google Scholar] [CrossRef] [Green Version]
- Shukla, D.; Liu, J.; Blaiklock, P.; Shworak, N.W.; Bai, X.; Esko, J.D.; Cohen, G.H.; Eisenberg, R.J.; Rosenberg, R.D.; Spear, P.G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 1999, 99, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Karasneh, G.A.; Shukla, D. Herpes simplex virus infects most cell types in vitro: Clues to its success. Virol. J. 2011, 8, 481. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.E.; Murray, J.W.; Wolkoff, A.W.; Wilson, D.W. Reconstitution of herpes simplex virus microtubule-dependent trafficking in vitro. J. Virol. 2006, 80, 4264–4275. [Google Scholar] [CrossRef] [Green Version]
- Lycke, E. Biological and molecular aspects on herpes simplex virus latency. Scand. J. Infect. Dis. Suppl. 1990, 69, 113–119. [Google Scholar]
- Hancock, M.H.; Corcoran, J.A.; Smiley, J.R. Herpes simplex virus regulatory proteins VP16 and ICP0 counteract an innate intranuclear barrier to viral gene expression. Virology 2006, 352, 237–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGeoch, D.J.; Rixon, F.J.; Davison, A.J. Topics in herpesvirus genomics and evolution. Virus Res. 2006, 117, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Steiner, I.; Kennedy, P.G.E. Herpes simplex virus latency in the nervous system—A new model. Neuropathol. Appl. Neurobiol. 1991, 17, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D. ICPO, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 2000, 22, 761–770. [Google Scholar] [CrossRef]
- Hadigal, S.R.; Agelidis, A.M.; Karasneh, G.A.; Antoine, T.E.; Yakoub, A.M.; Ramani, V.C.; Djalilian, A.R.; Sanderson, R.D.; Shukla, D. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat. Commun. 2015, 6, 6985. [Google Scholar] [CrossRef] [Green Version]
- Moore, M.D.; Bunka, D.H.J.; Forzan, M.; Spear, P.G.; Stockley, P.G.; Mcgowan, I.; James, W. Generation of neutralizing aptamers against herpes simplex virus type 2: Potential components of multivalent microbicides. J. Gen. Virol. 2011, 92, 1493–1499. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Hayashi, K.; Kumar, P.K.R. Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J. Virol. 2012, 86, 6732–6744. [Google Scholar] [CrossRef] [Green Version]
- Yadavalli, T.; Agelidis, A.; Jaishankar, D.; Mangano, K.; Thakkar, N.; Penmetcha, K.; Shukla, D. Targeting herpes simplex virus-1 gD by a DNA aptamer can be an effective new strategy to curb viral infection. Mol. Ther. Nucleic Acids 2017, 9, 365–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, W.; Spivak, D.A. A double-imprinted diffraction-grating sensor based on a virus-responsive super-aptamer hydrogel derived from an impure extract. Angew. Chem. Int. Ed. 2014, 53, 2095–2098. [Google Scholar] [CrossRef]
- Park, J.-W.; Lee, S.J.; Choi, E.-J.; Kim, J.; Song, J.-Y.; Gu, M.B. An ultra-sensitive detection of a whole virus using dual aptamers developed by immobilization-free screening. Biosens. Bioelectron. 2014, 51, 324–329. [Google Scholar] [CrossRef]
- Roh, C.; Jo, S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011, 86, 1475–1479. [Google Scholar] [CrossRef]
- Moore, M.D.; Escudero-Abarca, B.I.; Suh, S.H.; Jaykus, L.-A. Generation and characterization of nucleic acid aptamers targeting the capsid P domain of a human norovirus GII. 4 strain. J. Biotechnol. 2015, 209, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escudero-Abarca, B.I.; Suh, S.H.; Moore, M.D.; Dwivedi, H.P.; Jaykus, L.-A. Selection, characterization and application of nucleic acid aptamers for the capture and detection of human norovirus strains. PLoS ONE 2014, 9, e106805. [Google Scholar] [CrossRef] [PubMed]
- Giamberardino, A.; Labib, M.; Hassan, E.M.; Tetro, J.A.; Springthorpe, S.; Sattar, S.A.; Berezovski, M.V.; DeRosa, M.C. Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS ONE 2013, 8, e79087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.J.; Kwon, Y.S.; Lee, J.-E.; Choi, E.-J.; Lee, C.-H.; Song, J.-Y.; Gu, M.B. Detection of VR-2332 strain of porcine reproductive and respiratory syndrome virus type II using an aptamer-based sandwich-type assay. Anal. Chem. 2013, 85, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Zamay, A.S.; Berezovski, M.V. Multifunctional electrochemical aptasensor for aptamer clones screening, virus quantitation in blood and viability assessment. Analyst 2013, 138, 1865–1875. [Google Scholar] [CrossRef]
- Labib, M.; Zamay, A.S.; Muharemagic, D.; Chechik, A.V.; Bell, J.C.; Berezovski, M.V. Aptamer-based viability impedimetric sensor for viruses. Anal. Chem. 2012, 84, 1813–1816. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Zamay, A.S.; Muharemagic, D.; Chechik, A.; Bell, J.C.; Berezovski, M.V. Electrochemical sensing of aptamer-facilitated virus immunoshielding. Anal. Chem. 2012, 84, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Wehbe, M.; Labib, M.; Muharemagic, D.; Zamay, A.S.; Berezovski, M.V. Switchable aptamers for biosensing and bioseparation of viruses (SwAps-V). Biosens. Bioelectron. 2015, 67, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Komorowska, B.; Hasiów-Jaroszewska, B.; Minicka, J. Application of nucleic acid aptamers for detection of Apple stem pitting virus isolates. Mol. Cell. Probes 2017, 36, 62–65. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Zhou, S.; Shen, J.; Yang, D.; Wu, J.; Li, X.; Li, M.; Huang, X.; Sealy, J.E.; et al. A DNA aptamer efficiently inhibits the infectivity of Bovine herpesvirus 1 by blocking viral entry. Sci. Rep. 2017, 7, 11796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cnossen, E.J.; Silva, A.G.; Marangoni, K.; Arruda, R.A.; Souza, E.G.; Santos, F.A.; Fujimura, P.T.; Yokosawa, J.; Goulart, L.R.; Neves, A.F. Characterization of oligonucleotide aptamers targeting the 5′-UTR from dengue virus. Future Med. Chem. 2017, 9, 541–552. [Google Scholar] [CrossRef]
- Trausch, J.J.; Shank-Retzlaff, M.; Verch, T. Development and characterization of an HPV type-16 specific modified DNA aptamer for the improvement of potency assays. Anal. Chem. 2017, 89, 3554–3561. [Google Scholar] [CrossRef]
- Li, P.; Zhou, L.; Wei, J.; Yu, Y.; Yang, M.; Wei, S.; Qin, Q. Development and characterization of aptamer-based enzyme-linked apta-sorbent assay for the detection of Singapore grouper iridovirus infection. J. Appl. Microbiol. 2016, 121, 634–643. [Google Scholar] [CrossRef]
- Lu, T.; Ma, Q.; Yan, W.; Wang, Y.; Zhang, Y.; Zhao, L.; Chen, H. Selection of an aptamer against Muscovy duck parvovirus for highly sensitive rapid visual detection by label-free aptasensor. Talanta 2018, 176, 214–220. [Google Scholar] [CrossRef]
- Percze, K.; Szakács, Z.; Scholz, É.; András, J.; Szeitner, Z.; van den Kieboom, C.H.; Ferwerda, G.; de Jonge, M.I.; Gyurcsányi, R.E.; Mészáros, T. Aptamers for respiratory syncytial virus detection. Sci. Rep. 2017, 7, srep42794. [Google Scholar] [CrossRef] [PubMed]
- Nilaratanakul, V.; Hauer, D.A.; Griffin, D.E. Development and characterization of sindbis virus with encoded fluorescent RNA aptamer Spinach2 for imaging of replication and immune-mediated changes in intracellular viral RNA. J. Gen. Virol. 2017, 98, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.L.; Yu, D.; Smolke, C.D.; Geall, A.J.; Beard, C.W.; Mason, P.W. Control of alphavirus-based gene expression using engineered riboswitches. Virology 2015, 483, 302–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.-J.; Woo, H.-M.; Kim, K.-S.; Oh, J.-W.; Jeong, Y.-J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011, 112, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-L.; Hsiao, W.-H.; Lee, H.-C.; Wu, S.-C.; Cheng, J.-W. Selection and characterization of DNA aptamers targeting all four serotypes of dengue viruses. PLoS ONE 2015, 10, e0131240. [Google Scholar] [CrossRef] [PubMed]
- Gandham, S.H.A.; Volk, D.E.; Lokesh, G.L.R.; Neerathilingam, M.; Gorenstein, D.G. Thioaptamers targeting dengue virus type-2 envelope protein domain III. Biochem. Biophys. Res. Commun. 2014, 453, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Binning, J.M.; Wang, T.; Luthra, P.; Shabman, R.S.; Borek, D.M.; Liu, G.; Xu, W.; Leung, D.W.; Basler, C.F.; Amarasinghe, G.K. Development of RNA aptamers targeting Ebola virus VP35. Biochemistry 2013, 52, 8406–8419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrest, S.; Lear, Z.; Herod, M.R.; Ryan, M.; Rowlands, D.J.; Stonehouse, N.J. Inhibition of the foot-and-mouth disease virus subgenomic replicon by RNA aptamers. J. Gen. Virol. 2014, 95, 2649–2657. [Google Scholar] [CrossRef] [Green Version]
- Punnarak, P.; Santos, M.D.; Hwang, S.D.; Kondo, H.; Hirono, I.; Kikuchi, Y.; Aoki, T. RNA aptamers inhibit the growth of the fish pathogen viral hemorrhagic septicemia virus (VHSV). Mar. Biotechnol. 2012, 14, 752–761. [Google Scholar] [CrossRef]
- Hwang, S.D.; Midorikawa, N.; Punnarak, P.; Kikuchi, Y.; Kondo, H.; Hirono, I.; Aoki, T. Inhibition of hirame rhabdovirus growth by RNA aptamers. J. Fish Dis. 2012, 35, 927–934. [Google Scholar] [CrossRef]
- Toscano-Garibay, J.D.; Benítez-Hess, M.L.; Alvarez-Salas, L.M. Targeting of the HPV-16 E7 protein by RNA aptamers. Methods Mol. Biol. 2015, 1249, 221–239. [Google Scholar] [CrossRef]
- Leija-Montoya, A.G.; Benítez-Hess, M.L.; Toscano-Garibay, J.D.; Alvarez-Salas, L.M. Characterization of an RNA aptamer against HPV-16 L1 virus-like particles. Nucleic Acid Ther. 2014, 24, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Gourronc, F.A.; Rockey, W.M.; Thiel, W.H.; Giangrande, P.H.; Klingelhutz, A.J. Identification of RNA aptamers that internalize into HPV-16 E6/E7 transformed tonsillar epithelial cells. Virology 2013, 446, 325–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Wei, S.; Zhou, L.; Yang, M.; Yu, Y.; Wei, J.; Jiang, G.; Qin, Q. Selection and characterization of novel DNA aptamers specifically recognized by Singapore grouper iridovirus-infected fish cells. J. Gen. Virol. 2015, 96, 3348–3359. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yan, Y.; Wei, S.; Wei, J.; Gao, R.; Huang, X.; Huang, Y.; Jiang, G.; Qin, Q. Isolation and characterization of a new class of DNA aptamers specific binding to Singapore grouper iridovirus (SGIV) with antiviral activities. Virus Res. 2014, 188, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Beier, R.; Pahlke, C.; Quenzel, P.; Henseleit, A.; Boschke, E.; Cuniberti, G.; Labudde, D. Selection of a DNA aptamer against norovirus capsid protein VP1. FEMS Microbiol. Lett. 2014, 351, 162–169. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.-R.; Hu, G.-Q.; Xue, X.-H.; Li, L.; Zheng, X.-X.; Gao, Y.-W.; Yang, S.-T.; Xia, X.-Z. Selection of an aptamer against rabies virus: A new class of molecules with antiviral activity. Virus Res. 2014, 184, 7–13. [Google Scholar] [CrossRef]
- Liang, H.-R.; Hu, G.-Q.; Li, L.; Gao, Y.-W.; Yang, S.-T.; Xia, X.-Z. Aptamers targeting rabies virus-infected cells inhibit street rabies virus in vivo. Int. Immunopharmacol. 2014, 21, 432–438. [Google Scholar] [CrossRef]
- Liang, H.-R.; Liu, Q.; Zheng, X.-X.; Gai, W.-W.; Xue, X.-H.; Hu, G.-Q.; Wu, H.-X.; Wang, H.-L.; Yang, S.-T.; Xia, X.-Z. Aptamers targeting rabies virus-infected cells inhibit viral replication both in vitro and in vivo. Virus Res 2013, 173, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.-R.; Hu, G.-Q.; Zhang, T.; Yang, Y.-J.; Zhao, L.-L.; Qi, Y.-L.; Wang, H.-L.; Gao, Y.-W.; Yang, S.-T.; Xia, X.-Z. Isolation of ssDNA aptamers that inhibit rabies virus. Int. Immunopharmacol. 2012, 14, 341–347. [Google Scholar] [CrossRef]
- Ellenbecker, M.; Sears, L.; Li, P.; Lanchy, J.-M.; Lodmell, J.S. Characterization of RNA aptamers directed against the nucleocapsid protein of rift valley fever virus. Antivir. Res. 2012, 93, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndongwe, T.P.; Adedeji, A.O.; Michailidis, E.; Ong, Y.T.; Hachiya, A.; Marchand, B.; Ryan, E.M.; Rai, D.K.; Kirby, K.A.; Whatley, A.S.; et al. Biochemical, inhibition and inhibitor resistance studies of xenotropic murine leukemia virus-related virus reverse transcriptase. Nucleic Acids Res. 2012, 40, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Schütze, T.; Wilhelm, B.; Greiner, N.; Braun, H.; Peter, F.; Mörl, M.; Erdmann, V.A.; Lehrach, H.; Konthur, Z.; Menger, M.; et al. Probing the SELEX process with next-generation sequencing. PLoS ONE 2011, 6, e29604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eulberg, D.; Buchner, K.; Maasch, C.; Klussmann, S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: Identification of a biostable substance P antagonist. Nucleic Acids Res. 2005, 33, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blind, M.; Blank, M. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids 2015, 4, e223. [Google Scholar] [CrossRef] [PubMed]
- Sabri, M.Z.; Hamid, A.A.A.; Hitam, S.M.S.; Rahim, M.Z.A. In-silico selection of aptamer: A review on the revolutionary approach to understand the aptamer design and interaction through computational chemistry. Mater. Today Proc. 2019, 19, 1572–1581. [Google Scholar] [CrossRef]
- Chushak, Y.; Stone, M.O. In silico selection of RNA aptamers. Nucleic Acids Res. 2009, 37, e87. [Google Scholar] [CrossRef] [Green Version]
- Saliva-Based COVID-19 DNA Aptamer Test: Formative Usability and Internal Validation Study. Available online: https://clinicaltrials.gov/ct2/show/NCT04974203 (accessed on 20 August 2021).
- Yang, G.; Li, Z.; Mohammed, I.; Zhao, L.; Wei, W.; Xiao, H.; Guo, W.; Zhao, Y.; Qu, F.; Huang, Y. Identification of SARS-CoV-2-against aptamer with high neutralization activity by blocking the RBD domain of spike protein 1. Signal Transduct. Target Ther. 2021, 6, 227. [Google Scholar] [CrossRef]
- Schmitz, A.; Weber, A.; Bayin, M.; Breuers, S.; Fieberg, V.; Famulok, M.; Mayer, G. A SARS-CoV-2 spike binding DNA aptamer that inhibits pseudovirus infection by an RBD-independent mechanism. Angew. Chem. Int. Ed. 2021, 60, 10279–10285. [Google Scholar] [CrossRef]
- Pramanik, A.; Gao, Y.; Patibandla, S.; Mitra, D.; McCandless, M.G.; Fassero, L.A.; Gates, K.; Tandon, R.; Ray, P.C. Aptamer conjugated gold nanostar-based distance-dependent nanoparticle surface energy transfer spectroscopy for ultrasensitive detection and inactivation of corona virus. J. Phys. Chem. Lett. 2021, 12, 2166–2171. [Google Scholar] [CrossRef]
- Ando, T.; Yamamoto, M.; Takamori, Y.; Tsukamoto, K.; Fuji, D.; Kawakami, T. In vitro selection of an RNA aptamer yields an interleukin-6/interleukin-6 receptor interaction inhibitor. Biosci. Biotechnol. Biochem. 2021, 85, 1170–1174. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadavalli, T.; Volety, I.; Shukla, D. Aptamers in Virology—A Consolidated Review of the Most Recent Advancements in Diagnosis and Therapy. Pharmaceutics 2021, 13, 1646. https://doi.org/10.3390/pharmaceutics13101646
Yadavalli T, Volety I, Shukla D. Aptamers in Virology—A Consolidated Review of the Most Recent Advancements in Diagnosis and Therapy. Pharmaceutics. 2021; 13(10):1646. https://doi.org/10.3390/pharmaceutics13101646
Chicago/Turabian StyleYadavalli, Tejabhiram, Ipsita Volety, and Deepak Shukla. 2021. "Aptamers in Virology—A Consolidated Review of the Most Recent Advancements in Diagnosis and Therapy" Pharmaceutics 13, no. 10: 1646. https://doi.org/10.3390/pharmaceutics13101646
APA StyleYadavalli, T., Volety, I., & Shukla, D. (2021). Aptamers in Virology—A Consolidated Review of the Most Recent Advancements in Diagnosis and Therapy. Pharmaceutics, 13(10), 1646. https://doi.org/10.3390/pharmaceutics13101646