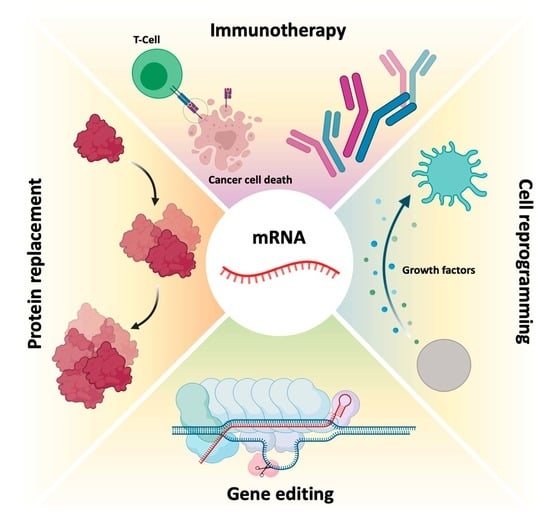
mRNA, a Revolution in Biomedicine
Abstract
:1. Introduction
2. mRNA Structure and Biogenesis
3. Therapeutic Application: mRNA vs. DNA
4. Production of mRNA
5. Challenges in mRNA Delivery
6. mRNA Therapies
6.1. Protein Replacement Therapies
6.2. Immunotherapy
6.3. Gene Editing
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2. [Google Scholar] [CrossRef]
- Pierce, B. Genetics: A Conceptual Approach, 6th ed.; W.H. Freeman and Company: New York, NY, USA, 2017. [Google Scholar]
- Nguyen, T.C.; Zaleta-Rivera, K.; Huang, X.; Dai, X.; Zhong, S. RNA, Action through Interactions. Trends Genet. 2018, 34, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Saldi, T.; Riemondy, K.; Erickson, B.; Bentley, D.L. Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Mol. Cell 2021, 81, 1789–1801.e5. [Google Scholar] [CrossRef]
- Lehman, N. RNA in evolution. Wiley Interdiscip. Rev. RNA 2010, 1, 202–213. [Google Scholar] [CrossRef]
- Chujo, T.; Yamazaki, T.; Hirose, T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim. Biophys. Acta (BBA)—Bioenerg. 2016, 1859, 139–146. [Google Scholar] [CrossRef]
- Bicknell, A.A.; Ricci, E.P. When mRNA translation meets decay. Biochem. Soc. Trans. 2017, 45, 339–351. [Google Scholar] [CrossRef]
- Kowalski, P.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [Green Version]
- Kornberg, R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 12955–12961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Detke, S.; Stein, J.; Stein, G. Synthesis of histone messenger RNAs by RNA polymerase II in nuclei from S phase HeLa S3cells. Nucleic Acids Res. 1978, 5, 1515–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlake, T.; Thess, A.; Thran, M.; Jordan, I. mRNA as novel technology for passive immunotherapy. Cell. Mol. Life Sci. 2018, 76, 301–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Mc Cafferty, S.; Combes, F.; Huysmans, H.; De Temmerman, J.; Gitsels, A.; Vanrompay, D.; Catani, J.P.; Sanders, N.N. mRNA therapeutics deliver a hopeful message. Nano Today 2018, 23, 16–39. [Google Scholar] [CrossRef]
- Granot-Matok, Y.; Kon, E.; Dammes, N.; Mechtinger, G.; Peer, D. Therapeutic mRNA delivery to leukocytes. J. Control. Release 2019, 305, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Mao, Y.; Ji, Q.; Dersh, D.; Yewdell, J.W.; Qian, S.-B. Decoding mRNA translatability and stability from the 5′ UTR. Nat. Struct. Mol. Biol. 2020, 27, 1–8. [Google Scholar] [CrossRef]
- Trepotec, Z.; Lichtenegger, E.; Plank, C.; Aneja, M.K.; Rudolph, C. Delivery of mRNA Therapeutics for the Treatment of Hepatic Diseases. Mol. Ther. 2019, 27, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Magadum, A.; Kaur, K.; Zangi, L. mRNA-Based Protein Replacement Therapy for the Heart. Mol. Ther. 2019, 27, 785–793. [Google Scholar] [CrossRef] [Green Version]
- Sahu, I.; Haque, A.K.M.A.; Weidensee, B.; Weinmann, P.; Kormann, M.S.D. Recent Developments in mRNA-Based Protein Supplementation Therapy to Target Lung Diseases. Mol. Ther. 2019, 27, 803–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grozhik, A.V.; Jaffrey, S.R. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat. Chem. Biol. 2018, 14, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Eichhorn, S.W.; Subtelny, A.O.; Kronja, I.; Kwasnieski, J.C.; Orr-Weaver, T.L.; Bartel, D.P. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 2016, 5, e16955. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, K.; Passmore, L.A. Cytoplasmic deadenylation: Regulation of mRNA fate. Biochem. Soc. Trans. 2010, 38, 1531–1536. [Google Scholar] [CrossRef] [Green Version]
- Dreyfus, M.; Régnier, P. The poly (A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell 2002, 111, 611–613. [Google Scholar] [CrossRef] [Green Version]
- Schlake, T.; Thran, M.; Fiedler, K.; Heidenreich, R.; Petsch, B.; Fotin-Mleczek, M. mRNA: A Novel Avenue to Antibody Therapy? Mol. Ther. 2019, 27, 773–784. [Google Scholar] [CrossRef] [Green Version]
- Weng, Y.; Li, C.; Yang, T.; Hu, B.; Zhang, M.; Guo, S.; Xiao, H.; Liang, X.-J.; Huang, Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020, 40, 107534. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Perche, F.; Ikegami, M.; Uchida, S.; Kataoka, K.; Itaka, K. Messenger RNA-based therapeutics for brain diseases: An animal study for augmenting clearance of beta-amyloid by intracerebral administration of neprilysin mRNA loaded in polyplex nanomicelles. J. Control. Release 2016, 235, 268–275. [Google Scholar] [CrossRef]
- Guan, S.; Rosenecker, J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017, 24, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Park, S.-J.; Yim, Y.; Kim, J.; Choi, C.; Won, C.; Min, D.-H. Recent Advances in RNA Therapeutics and RNA Delivery Systems Based on Nanoparticles. Adv. Ther. 2018, 1. [Google Scholar] [CrossRef]
- Cao, J.; An, D.; Galduroz, M.; Zhuo, J.; Liang, S.; Eybye, M.; Frassetto, A.; Kuroda, E.; Funahashi, A.; Santana, J.; et al. mRNA Therapy Improves Metabolic and Behavioral Abnormalities in a Murine Model of Citrin Deficiency. Mol. Ther. 2019, 27, 1242–1251. [Google Scholar] [CrossRef] [Green Version]
- Midoux, P.; Pichon, C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccines 2014, 14, 221–234. [Google Scholar] [CrossRef] [Green Version]
- An, D.; Frassetto, A.; Jacquinet, E.; Eybye, M.; Milano, J.; DeAntonis, C.; Nguyen, V.; Laureano, R.; Milton, J.; Sabnis, S.; et al. Long-term efficacy and safety of mRNA therapy in two murine models of methylmalonic acidemia. EBioMedicine 2019, 45, 519–528. [Google Scholar] [CrossRef] [Green Version]
- Schlake, T.; Thess, A.; Fotin-Mleczek, M.; Kallen, K.-J. Developing mRNA-vaccine technologies. RNA Biol. 2012, 9, 1319–1330. [Google Scholar] [CrossRef] [Green Version]
- Karikó, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.A. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, A.-M.; Batra, N.; Tu, M.-J.; Sweeney, C. Novel approaches for efficient in vivo fermentation production of noncoding RNAs. Appl. Microbiol. Biotechnol. 2020, 104, 1927–1937. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.; Pedro, A.Q.; Queiroz, J.A.; Figueiras, A.R.; Sousa, F. New insights for therapeutic recombinant human miRNAs heterologous production: Rhodovolum sulfidophilum vs. Escherichia coli. Bioengineered 2017, 8, 670–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, P.; Pedro, A.Q.; Tomás, J.; Maia, C.J.; Queiroz, J.A.; Figueiras, A.; Sousa, F. Advances in time course extracellular production of human pre-miR-29b from Rhodovulum sulfidophilum. Appl. Microbiol. Biotechnol. 2016, 100, 3723–3734. [Google Scholar] [CrossRef] [PubMed]
- Baptista, B.; Riscado, M.; Queiroz, J.; Pichon, C.; Sousa, F. Non-coding RNAs: Emerging from the discovery to therapeutic applications. Biochem. Pharmacol. 2021, 189, 114469. [Google Scholar] [CrossRef]
- Roy, I.; Stachowiak, M.K.; Bergey, E.J. Nonviral gene transfection nanoparticles: Function and applications in the brain. Nanomed. Nanotechnol. Biol. Med. 2008, 4, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.A.; Xu, Y.; Tao, W.; Ubellacker, J.M.; Lim, M.; Aum, D.; Lee, G.Y.; Zhou, K.; Zope, H.; Yu, M.; et al. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2018, 2, 850–864. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorth, M.; Narvekar, A. Non viral vectors in gene therapy- an overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. [Google Scholar] [CrossRef]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef]
- Gómez-Aguado, I.; Rodríguez-Castejón, J.; Vicente-Pascual, M.; Rodríguez-Gascón, A.; Aspiazu, M.; Ángeles, S.; Del Pozo-Rodríguez, A. Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials 2020, 10, 364. [Google Scholar] [CrossRef] [Green Version]
- Rozovics, J.M.; Chase, A.J.; Cathcart, A.L.; Chou, W.; Gershon, P.D.; Palusa, S.; Wilusz, J.; Semler, B.L. Picornavirus Modification of a Host mRNA Decay Protein. mBio 2012, 3, e00431-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, S.; Yang, S.; Hu, X.; Yin, D.; Dai, Y.; Qian, X.; Wang, D.; Pan, X.; Hong, J.; Sun, X.; et al. Lentiviral delivery of co-packaged Cas9 mRNA and a Vegfa-targeting guide RNA prevents wet age-related macular degeneration in mice. Nat. Biomed. Eng. 2021, 5, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and Challenges in the Delivery of mRNA-Based Vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liang, H.; Liu, J.; Wang, Z. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int. J. Pharm. 2018, 546, 215–225. [Google Scholar] [CrossRef]
- Kaur, I.P.; Sharma, G.; Singh, M.; Sandhu, S.K.; Deol, P.; Yadav, M.; Yakhmi, J.V. Nanobiomaterials as gene-delivery vehicles. Nanobiomater. Drug Deliv. 2016, 9, 447–486. [Google Scholar] [CrossRef]
- Hassett, K.J.; Higgins, J.; Woods, A.; Levy, B.; Xia, Y.; Hsiao, C.J.; Acosta, E.; Almarsson, O.; Moore, M.J.; Brito, L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control. Release 2021, 335, 237–246. [Google Scholar] [CrossRef]
- Godbey, W.T.; Wu, K.K.; Mikos, A.G. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J. Biomed. Mater. Res. 1999, 45, 268–275. [Google Scholar] [CrossRef]
- Forrest, M.L.; Gabrielson, N.; Pack, D.W. Cyclodextrin-polyethylenimine conjugates for targeted in vitro gene delivery. Biotechnol. Bioeng. 2004, 89, 416–423. [Google Scholar] [CrossRef]
- Sultana, N.; Magadum, A.; Hadas, Y.; Kondrat, J.; Singh, N.; Youssef, E.; Calderon, D.; Chepurko, E.; Dubois, N.; Hajjar, R.J.; et al. Optimizing Cardiac Delivery of Modified mRNA. Mol. Ther. 2017, 25, 1306–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggli, N.; Demoulins, T.; Pichon, C.; Suter, R.; Guzmán, C.A.; Ebensen, T.; Midoux, P.; McCullough, K.; Milona, P.; Schulze, K.; et al. Polyethylenimine-based polyplex delivery of self-replicating RNA vaccines. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 711–722. [Google Scholar] [CrossRef]
- Zhao, M.; Li, M.; Zhang, Z.; Gong, T.; Sun, X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 2015, 23, 2596–2607. [Google Scholar] [CrossRef]
- Lallana, E.; de la Rosa, J.M.R.; Tirella, A.; Pelliccia, M.; Gennari, A.; Stratford, I.J.; Puri, S.; Ashford, M.; Tirelli, N. Chitosan/Hyaluronic Acid Nanoparticles: Rational Design Revisited for RNA Delivery. Mol. Pharm. 2017, 14, 2422–2436. [Google Scholar] [CrossRef]
- McCullough, K.C.; Bassi, I.; Milona, P.; Suter, R.; Thomann-Harwood, L.; Englezou, P.; Démoulins, T.; Ruggli, N. Self-replicating Replicon-RNA Delivery to Dendritic Cells by Chitosan-nanoparticles for Translation In Vitro and In Vivo. Mol. Ther.—Nucleic Acids 2014, 3, e173. [Google Scholar] [CrossRef]
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Goel, V.; Robbie, G.J. Pharmacokinetics of Patisiran, the First Approved RNA Interference Therapy in Patients With Hereditary Transthyretin-Mediated Amyloidosis. J. Clin. Pharmacol. 2019, 60, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Gerhardt, A.; Voigt, E.; Archer, M.; Reed, S.; Larson, E.; Van Hoeven, N.; Kramer, R.; Fox, C.; Casper, C. A Thermostable, Flexible RNA Vaccine Delivery Platform for Pandemic Response. bioRxiv 2021. [Google Scholar] [CrossRef]
- Malone, R.W.; Felgner, P.L.; Verma, I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA 1989, 86, 6077–6081. [Google Scholar] [CrossRef] [Green Version]
- Sayour, E.J.; De Leon, G.; Pham, C.; Grippin, A.; Kemeny, H.; Chua, J.; Huang, J.; Sampson, J.; Sanchez-Perez, L.; Flores, C.; et al. Systemic activation of antigen-presenting cells via RNA-loaded nanoparticles. OncoImmunology 2016, 6, e1256527. [Google Scholar] [CrossRef] [Green Version]
- Verbeke, R.; Lentacker, I.; Wayteck, L.; Breckpot, K.; Van Bockstal, M.; Descamps, B.; Vanhove, C.; De Smedt, S.C.; Dewitte, H. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: Restoring the immunogenicity of immunosilent mRNA. J. Control Release 2017, 266, 287–300. [Google Scholar] [CrossRef]
- Basha, G.; Novobrantseva, T.I.; Rosin, N.; Tam, Y.Y.C.; Hafez, I.M.; Wong, M.K.; Sugo, T.; Ruda, V.M.; Qin, J.; Klebanov, B.; et al. Influence of Cationic Lipid Composition on Gene Silencing Properties of Lipid Nanoparticle Formulations of siRNA in Antigen-Presenting Cells. Mol. Ther. 2011, 19, 2186–2200. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Sedic, M.; Senn, J.J.; Lynn, A.; Laska, M.; Smith, M.; Platz, S.J.; Bolen, J.; Hoge, S.; Bulychev, A.; Jacquinet, E.; et al. Safety Evaluation of Lipid Nanoparticle–Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2017, 55, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Arteta, M.Y.; Kjellman, T.; Bartesaghi, S.; Wallin, S.; Wu, X.; Kvist, A.J.; Dabkowska, A.; Székely, N.; Radulescu, A.; Bergenholtz, J.; et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA 2018, 115, E3351–E3360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardi, N.; Hogan, M.; Pelc, R.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Hekele, A.; Bertholet, S.; Archer, J.; Gibson, D.G.; Palladino, G.; Brito, L.A.; Otten, G.R.; Brazzoli, M.; Buccato, S.; Bonci, A.; et al. Rapidly produced SAM® vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013, 2, 1–7. [Google Scholar] [CrossRef]
- Guevara, M.L.; Persano, F.; Persano, S. Advances in lipid nanoparticles for mRNA-based cancer immunotherapy. Front. Chem. 2020, 8, 963. [Google Scholar] [CrossRef]
- Mockey, M.; Bourseau, E.; Chandrashekhar, V.; Chaudhuri, A.; Lafosse, S.; Le Cam, E.; Quesniaux, V.F.J.; Ryffel, B.; Pichon, C.; Midoux, P. mRNA-based cancer vaccine: Prevention of B16 melanoma progression and metastasis by systemic injection of MART1 mRNA histidylated lipopolyplexes. Cancer Gene Ther. 2007, 14, 802–814. [Google Scholar] [CrossRef] [Green Version]
- Petsch, B.; Schnee, M.; Vogel, A.B.; Lange, E.; Hoffmann, B.; Voss, D.; Schlake, T.; Thess, A.; Kallen, K.-J.; Stitz, L.; et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012, 30, 1210–1216. [Google Scholar] [CrossRef]
- Kübler, H.; Scheel, B.; Gnad-Vogt, U.; Miller, K.; Schultze-Seemann, W.; Dorp, F.V.; Parmiani, G.; Hampel, C.; Wedel, S.; Trojan, L.; et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: A first-in-man phase I/IIa study. J. Immunother. Cancer 2015, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Sebastian, M.; Schröder, A.; Scheel, B.; Hong, H.S.; Muth, A.; von Boehmer, L.; Zippelius, A.; Mayer, F.; Reck, M.; Atanackovic, D.; et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol. Immunother. 2019, 68, 799–812. [Google Scholar] [CrossRef]
- Armbruster, N.; Jasny, E.; Petsch, B. Advances in RNA Vaccines for Preventive Indications: A Case Study of a Vaccine Against Rabies. Vaccines 2019, 7, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalil, A.S.; Yu, X.; Umhoefer, J.M.; Chamberlain, C.S.; Wildenauer, L.A.; Diarra, G.M.; Hacker, T.A.; Murphy, W.L. Single-dose mRNA therapy via biomaterial-mediated sequestration of overexpressed proteins. Sci. Adv. 2020, 6, eaba2422. [Google Scholar] [CrossRef] [PubMed]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef]
- Mai, Y.; Guo, J.; Zhao, Y.; Ma, S.; Hou, Y.; Yang, J. Intranasal delivery of cationic liposome-protamine complex mRNA vaccine elicits effective anti-tumor immunity. Cell. Immunol. 2020, 354, 104143. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Tonnu, N.; Tachikawa, K.; Limphong, P.; Vega, J.B.; Karmali, P.P.; Chivukula, P.; Verma, I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. USA 2017, 114, E1941–E1950. [Google Scholar] [CrossRef] [Green Version]
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef]
- Baba, M.; Itaka, K.; Kondo, K.; Yamasoba, T.; Kataoka, K. Treatment of neurological disorders by introducing mRNA in vivo using polyplex nanomicelles. J. Control. Release 2015, 201, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Magadum, A.; Singh, N.; Kurian, A.A.; Sharkar, M.T.K.; Chepurko, E.; Zangi, L. Ablation of a Single N-Glycosylation Site in Human FSTL 1 Induces Cardiomyocyte Proliferation and Cardiac Regeneration. Mol. Ther.-Nucleic Acids 2018, 13, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Magadum, A.; Singh, N.; Kurian, A.A.; Munir, I.; Mehmood, T.; Brown, K.; Sharkar, M.T.K.; Chepurko, E.; Sassi, Y.; Oh, J.G.; et al. Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration. Circulation 2020, 141, 1249–1265. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Zhang, X.; Men, K.; Gao, Y.; Yang, X.; Wu, S.; Duan, X.; Wei, Y.; Tong, R. Efficient Colorectal Cancer Gene Therapy with IL-15 mRNA Nanoformulation. Mol. Pharm. 2020, 17. [Google Scholar] [CrossRef]
- Jiang, T.; Zhou, C.; Ren, S. Role of IL-2 in cancer immunotherapy. OncoImmunology 2016, 5, e1163462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, J.D.; Reidenbach, D.; Salomon, N.; Sahin, U.; Türeci, Ö.; Vormehr, M.; Kranz, L.M. mRNA therapeutics in cancer immunotherapy. Mol. Cancer 2021, 20, 69. [Google Scholar] [CrossRef]
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27, 757–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delehedde, C.; Even, L.; Midoux, P.; Pichon, C.; Perche, F. Intracellular Routing and Recognition of Lipid-Based mRNA Nanoparticles. Pharmaceutics 2021, 13, 945. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.-I.; Wu, C.-J.; Tu, Y.-F.; Chi, C.-Y. Fighting COVID-19: A quick review of diagnoses, therapies, and vaccines. Biomed. J. 2020, 43, 341–354. [Google Scholar] [CrossRef]
- Conry, R.M.; LoBuglio, A.F.; Wright, M.; Sumerel, L.; Pike, M.J.; Johanning, F.; Benjamin, R.; Lu, D.; Curiel, D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995, 55, 1397–1400. [Google Scholar]
- Billingsley, M.M.; Singh, N.; Ravikumar, P.; Zhang, R.; June, C.H.; Mitchell, M.J. Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett. 2020, 20, 1578–1589. [Google Scholar] [CrossRef]
- Belete, T.M. A review on Promising vaccine development progress for COVID-19 disease. Vacunas 2020, 21, 121–128. [Google Scholar] [CrossRef]
- He, W.; Evans, A.C.; Rasley, A.; Bourguet, F.; Peters, S.; Kamrud, K.I.; Wang, N.; Hubby, B.; Felderman, M.; Gouvis, H.; et al. Cationic HDL mimetics enhance in vivo delivery of self-replicating mRNA. Nanomed. Nanotechnol. Biol. Med. 2020, 24, 102154. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, L.; Almutairi, M.M.; Hotez, P.J.; Pollet, J. Enlisting the mRNA Vaccine Platform to Combat Parasitic Infections. Vaccines 2019, 7, 122. [Google Scholar] [CrossRef] [Green Version]
- Alton, E.W.F.W.; Armstrong, D.K.; Ashby, D.; Bayfield, K.J.; Bilton, D.; Bloomfield, E.V.; Boyd, A.C.; Brand, J.; Buchan, R.; Calcedo, R.; et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: A randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2015, 3, 684–691. [Google Scholar] [CrossRef] [Green Version]
- Alberer, M.; Gnad-Vogt, U.; Hong, H.S.; Mehr, K.T.; Backert, L.; Finak, G.; Gottardo, R.; Bica, M.A.; Garofano, A.; Koch, S.D.; et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: An open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet 2017, 390, 1511–1520. [Google Scholar] [CrossRef]
- Fotin-Mleczek, M.; Duchardt, K.M.; Lorenz, C.; Pfeiffer, R.; Ojkić-Zrna, S.; Probst, J.; Kallen, K.-J. Messenger RNA-based Vaccines with Dual Activity Induce Balanced TLR-7 Dependent Adaptive Immune Responses and Provide Antitumor Activity. J. Immunother. 2011, 34, 1–15. [Google Scholar] [CrossRef]
- Feldman, R.A.; Fuhr, R.; Smolenov, I.; Ribeiro, A.; Panther, L.; Watson, M.; Senn, J.J.; Smith, M.; Almarsson, Ӧ.; Pujar, H.S.; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334. [Google Scholar] [CrossRef]
- Bos, R.; Marquardt, K.L.; Cheung, J.; Sherman, L.A. Functional differences between low- and high-affinity CD8+ T cells in the tumor environment. OncoImmunology 2012, 1, 1239–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, L.; Hinterberger, M.; Wirnsberger, G.; Kyewski, B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 2009, 9, 833–844. [Google Scholar] [CrossRef]
- Leal, L.; Guardo, A.C.; Morón-López, S.; Salgado, M.; Mothe, B.; Heirman, C.; Pannus, P.; Vanham, G.; van den Ham, H.J.; Gruters, R.; et al. Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS 2018, 32, 2533–2545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jong, W.; Aerts, J.; Allard, S.; Brander, C.; Buyze, J.; Florence, E.; van Gorp, E.; Vanham, G.; Leal, L.; Mothe, B. iHIVARNA phase IIa, a randomized, placebo-controlled, double-blinded trial to evaluate the safety and immunogenicity of iHIVARNA-01 in chronically HIV-infected patients under stable combined antiretroviral therapy. Trials 2019, 20, 1–10. [Google Scholar]
- Gandhi, R.T.; Kwon, D.S.; Macklin, E.A.; Shopis, J.R.; McLean, A.P.; McBrine, N.; Flynn, T.; Peter, L.; Sbrolla, A.; Kaufmann, D.E. Immunization of HIV-1-infected persons with autologous dendritic cells transfected with mRNA encoding HIV-1 Gag and Nef: Results of a randomized, placebo-controlled clinical trial. J. Acquir. Immune Defic. Syndr. 2016, 71, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyte, J.A.; Mu, L.; Aamdal, S.; Kvalheim, G.; Dueland, S.; Hauser, M.; Gullestad, H.P.; Ryder, T.; Lislerud, K.; Hammerstad, H.; et al. Phase I/II trial of melanoma therapy with dendritic cells transfected with autologous tumor-mRNA. Cancer Gene Ther. 2006, 13, 905–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wecker, M.; Gilbert, P.; Russell, N.; Hural, J.; Allen, M.; Pensiero, M.; Chulay, J.; Chiu, Y.-L.; Karim, S.S.A.; Burke, D.S. Phase I Safety and Immunogenicity Evaluations of an Alphavirus Replicon HIV-1 Subtype C gag Vaccine in Healthy HIV-1-Uninfected Adults. Clin. Vaccine Immunol. 2012, 19, 1651–1660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Ding, W.; Zhu, D.; Yu, L.; Jiang, X.; Wang, X.; Zhang, C.; Wang, L.; Ji, T.; Liu, D.; et al. TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J. Clin. Investig. 2014, 125, 425–436. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Yu, L.; Zhu, D.; Ding, W.; Wang, X.; Zhang, C.; Wang, L.; Jiang, X.; Shen, H.; He, D.; et al. Disruption of HPV16-E7 by CRISPR/Cas System Induces Apoptosis and Growth Inhibition in HPV16 Positive Human Cervical Cancer Cells. BioMed Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Qasim, W.; Zhan, H.; Samarasinghe, S.; Adams, S.; Amrolia, P.; Stafford, S.; Butler, K.; Rivat, C.; Wright, G.; Somana, K.; et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017, 9, eaaj2013. [Google Scholar] [CrossRef]
- Su, S.; Hu, B.; Shao, J.; Shen, B.; Du, J.; Du, Y.; Zhou, J.; Yu, L.; Zhang, L.; Chen, F.; et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016, 6, 20070. [Google Scholar] [CrossRef] [Green Version]
- Quan, L.; Chen, X.; Liu, A.; Zhang, Y.; Guo, X.; Yan, S.; Liu, Y. PD-1 Blockade Can Restore Functions of T-Cells in Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma In Vitro. PLoS ONE 2015, 10, e0136476. [Google Scholar] [CrossRef]
- DiGiusto, D.L.; Cannon, P.M.; Holmes, M.C.; Li, L.; Rao, A.; Wang, J.; Lee, G.; Gregory, P.; Kim, K.A.; Hayward, S.B.; et al. Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Mol. Ther.—Methods Clin. Dev. 2016, 3, 16067. [Google Scholar] [CrossRef] [Green Version]
- Samson, M.; Libert, F.; Doranz, B.J.; Rucker, J.; Liesnard, C.; Farber, C.-M.; Saragosti, S.; Lapoumeroulie, C.; Cognaux, J.; Forceille, C.; et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 1996, 382, 722–725. [Google Scholar] [CrossRef]
- Ahammad, I.; Lira, S.S. Designing a novel mRNA vaccine against SARS-CoV-2: An immunoinformatics approach. Int. J. Biol. Macromol. 2020, 162, 820–837. [Google Scholar] [CrossRef]
- Khuroo, M.S.; Khuroo, M.; Khuroo, M.S.; Sofi, A.A.; Khuroo, N.S. COVID-19 Vaccines: A Race Against Time in the Middle of Death and Devastation! J. Clin. Exp. Hepatol. 2020, 10, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Velavan, T.P.; Meyer, C.G. The COVID-19 epidemic. Trop. Med. Int. Health 2020, 25, 278–280. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Wang, X. COVID-19: A new challenge for human beings. Cell. Mol. Immunol. 2020, 17, 555–557. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [Green Version]
- Koirala, A.; Joo, Y.J.; Khatami, A.; Chiu, C.; Britton, P.N. Vaccines for COVID-19: The current state of play. Paediatr. Respir. Rev. 2020, 35, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Caddy, S. Developing a vaccine for COVID-19. BMJ 2020, 369, m1790. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Shen, H.-M. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, Q.; Li, Y.; Garner, L.V.; Watkins, S.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S.; et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Lurie, N.; Saville, M.; Hatchett, R.; Halton, J. Developing COVID-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020, 382, 1969–1973. [Google Scholar] [CrossRef]
- Thanh Le, T.; Andreadakis, Z.; Kumar, A.; Gómez Román, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar] [CrossRef] [PubMed]
- Corey, B.L.; Mascola, J.R.; Fauci, A.S.; Collins, F.S. A strategic approach to COVID-19 vaccine R&D. Science 2020, 368, 948–950. [Google Scholar] [CrossRef]
- Kowalzik, F.; Schreiner, D.; Jensen, C.; Teschner, D.; Gehring, S.; Zepp, F. mRNA-Based Vaccines. Vaccines 2021, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- Oberhardt, V.; Luxenburger, H.; Kemming, J.; Schulien, I.; Ciminski, K.; Giese, S.; Csernalabics, B.; Lang-Meli, J.; Janowska, I.; Staniek, J.; et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature 2021, 597, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Kim, J.H.; Marks, F.; Clemens, J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021, 27, 205–211. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Comirnaty Epar Product Information. Available online: https://www.ema.europa.eu/en/documents/product-information/comirnaty-epar-product-information_en.pdf (accessed on 18 November 2021).
- Spikevax previously COVID-19 vaccine Moderna Epar Product Information. Available online: https://www.ema.europa.eu/en/documents/product-information/spikevax-previously-covid-19-vaccine-moderna-epar-product-information_en.pdf (accessed on 18 November 2021).
- Lamb, Y.N. BNT162b2 mRNA COVID-19 Vaccine: First Approval. Drugs 2021, 81, 495–501. [Google Scholar] [CrossRef]
- Fact Sheet for Healthcare Providers Administering Vaccine (Vaccination Providers)—Emergency Use Authorization (EUA) of the Pfizer-Biontech COVID-19 Vaccine to Prevent Coronavirus Disease 2019 (COVID-19). Available online: https://www.fda.gov/media/153713/download (accessed on 26 November 2021).
- Fact Sheet for Healthcare Providers Administering Vaccine. Moderna. U.S. Food Drug Adm. 2020, 8, 55. Available online: https://www.fda.gov/media/144637/download (accessed on 8 May 2021).
- Comirnaty (COVID-19 mRNA Vaccine [Nucleoside Modified])—An Overview of Comirnaty and Why It Is Authorised in the EU. Available online: https://www.ema.europa.eu/en/documents/overview/comirnaty-epar-medicine-overview_en.pdf (accessed on 26 November 2021).
- Spikevax (COVID-19 mRNA Vaccine [Nucleoside Modified])—An Overview of Spikevax and Why It Is Authorised in the EU. Available online: https://www.ema.europa.eu/en/documents/overview/spikevax-previously-covid-19-vaccine-moderna-epar-medicine-overview_en.pdf (accessed on 26 November 2021).
- Widge, A.T.; Rouphael, N.G.; Jackson, L.A.; Anderson, E.J.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2020, 384, 80–82. [Google Scholar] [CrossRef]
- Meo, S.A.; Bukhari, I.A.; Akram, J.; Meo, A.S.; Klonoff, D.C. COVID-19 vaccines: Comparison of biological, pharmacological characteristics and adverse effects of Pfizer/BioNTech and Moderna Vaccines. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1663–1669. [Google Scholar] [PubMed]
- Kremsner, P.G.; Mann, P.; Kroidl, A.; Leroux-Roels, I.; Schindler, C.; Gabor, J.J.; Schunk, M.; Leroux-Roels, G.; Bosch, J.J.; Fendel, R.; et al. Safety and immunogenicity of an mRNA-lipid nanoparticle vaccine candidate against SARS-CoV-2. Wien. Klin. Wochenschr. 2021, 133, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; Roth, N.; Schwendt, K.; Fotin-Mleczek, M.; Mueller, S.O.; Petsch, B. mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. npj Vaccines 2021, 6, 57. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 COVID-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef]
- Sheikh, A.; McMenamin, J.; Taylor, B.; Robertson, C.; Public Health Scotland and the EAVE II Collaborators. SARS-CoV-2 Delta VOC in Scotland: Demographics, risk of hospital admission, and vaccine effectiveness. Lancet 2021, 397, 2461–2462. [Google Scholar] [CrossRef]
- Oliver, S.E.; Gargano, J.W.; Marin, M.; Wallace, M.; Curran, K.G.; Chamberland, M.; McClung, N.; Campos-Outcalt, D.; Morgan, R.L.; Mbaeyi, S.; et al. The Advisory Committee on Immunization Practices’ Interim Recommendation for Use of Moderna COVID-19 Vaccine - United States, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2021, 69, 1653–1656. [Google Scholar] [CrossRef]
- Vaidyanathan, S.; Azizian, K.T.; Haque, A.A.; Henderson, J.M.; Hendel, A.; Shore, S.; Antony, J.S.; Hogrefe, R.I.; Kormann, M.S.; Porteus, M.H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther.—Nucleic Acids 2018, 12, 530–542. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector with Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Eyler, D.E.; Franco, M.K.; Batool, Z.; Wu, M.Z.; Dubuke, M.L.; Dobosz-Bartoszek, M.; Jones, J.; Polikanov, Y.S.; Roy, B.; Koutmou, K.S. Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. USA 2019, 116, 23068–23074. [Google Scholar] [CrossRef] [PubMed]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Alexaki, A.; Hettiarachchi, G.K.; Athey, J.C.; Katneni, U.; Simhadri, V.; Hamasaki-Katagiri, N.; Nanavaty, P.; Lin, B.; Takeda, K.; Freedberg, D.; et al. Effects of codon optimization on coagulation factor IX translation and structure: Implications for protein and gene therapies. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Türeci, O.; Sahin, U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef]
- OMS. COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 1 October 2021).
- Ghosh, D.; Venkataramani, P.; Nandi, S.; Bhattacharjee, S. CRISPR–Cas9 a boon or bane: The bumpy road ahead to cancer therapeutics. Cancer Cell Int. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Janssen, J.M.; Chen, X.; Liu, J.; Gonçalves, M.A. The Chromatin Structure of CRISPR-Cas9 Target DNA Controls the Balance between Mutagenic and Homology-Directed Gene-Editing Events. Mol. Ther.-Nucleic Acids 2019, 16, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Shen, B.; Zhang, W.; Zhang, J.; Zhou, J.; Wang, J.; Chen, L.; Wang, L.; Hodgkins, A.; Iyer, V.; Huang, X.; et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 2014, 11, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Zhang, S.; Kos, P.; Xiong, H.; Zhou, K.; Perelman, S.S.; Zhu, H.; Siegwart, D.J. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2017, 56, 1059–1063. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohsin, M.; Li, Y.; Zhang, X.; Wang, Y.; Huang, Z.; Yin, G.; Zhang, Z. Development of CRISPR-CAS9 based RNA drugs against Eimeria tenella infection. Genomics 2021, 113, 4126–4135. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics 2013, 194, 1029–1035. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, M.; Glass, Z.; Chen, J.; Haas, M.; Jin, X.; Zhao, X.; Rui, X.; Ye, Z.; Li, Y.; Zhang, F.; et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Graham, M.J.; Lee, R.G.; Brandt, T.A.; Tai, L.-J.; Fu, W.; Peralta, R.; Yu, R.; Hurh, E.; Paz, E.; McEvoy, B.W.; et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N. Engl. J. Med. 2017, 377, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865.e12–876.e12. [Google Scholar] [CrossRef]
- Guo, X.; Rahman, J.A.; Wessels, H.-H.; Méndez-Mancilla, A.; Haro, D.; Chen, X.; Sanjana, N.E. Transcriptome-wide Cas13 guide RNA design for model organisms and viral RNA pathogens. Cell Genom. 2021, 1, 100001. [Google Scholar] [CrossRef]
- Kushawah, G.; Hernandez-Huertas, L.; Abugattas-Nuñez Del Prado, J.; Martinez-Morales, J.R.; DeVore, M.L.; Hassan, H.; Moreno-Sanchez, I.; Tomas-Gallardo, L.; Diaz-Moscoso, A.; Monges, D.E.; et al. CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos. Dev. Cell 2020, 54, 805.e7–817.e7. [Google Scholar] [CrossRef]
- Xu, D.; Cai, Y.; Tang, L.; Han, X.; Gao, F.; Cao, H.; Qi, F.; Kapranov, P. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, E.L.; Vanover, D.; Bawage, S.S.; Tiwari, P.M.; Rotolo, L.; Beyersdorf, J.; Peck, H.E.; Bruno, N.C.; Hincapie, R.; Michel, F.; et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol. 2021, 39, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Rashnonejad, A.; Chermahini, G.A.; Wallace, L.; Harper, S.O. 8DUX4 mRNA silencing with CRISPR-Cas13 gene therapy as a prospective treatment for Facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2019, 29, S40. [Google Scholar] [CrossRef]









| Characteristics | DNA  | RNA  |
|---|---|---|
| Type of sugar | Deoxyribose | Ribose |
| Bases | A, T, C, G | A, U, C, G |
| Double or single stranded | Double | Single |
| Secondary structure | Double helix | Many types |
| Stability | Stable | Easily degraded |
| Therapeutic Approach | Objective/Function |
|---|---|
| Protein Replacement | Restore function, increase expression or replace protein in rare monogenic diseases |
| Cell reprogramming | Modulate cellular behavior by expressing transcription and/or growth factors |
| Immunotherapies | Elicit specific immune responses against target cells, for example through therapeutic antibodies |
| mRNA Vaccines | Structure | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Non-amplifying mRNA Vaccines | Basic structure of the mRNA, with a coding region for the desired antigens. | - Relatively small mRNA size (~2–3 kb). - Absence of additional proteins, minimizing unwanted immune interactions. - Relatively easy to produce and amplify. - Simplified sequence engineering. - Direct antigen expression. | - Potential toxicity from modified nucleotides. - Short duration of expression. - Need for high RNA doses. - Low antigen quantity. | [8,86] |
| SAM Vaccines | Encode a manipulated RNA virus genome (replicon). It generally contains two different protein coding regions, one encoding nonstructural proteins involved in mRNA capping and replication, and the other in antigen expression. | - High yield of target antigen. - Enhanced and prolonged antigen expression. - Lower effective RNA doses (more safe). - Intrinsic adjuvant effect. - Potential apoptosis of vaccine-carrying cells due to vaccine self-amplification (enhanced cross-presentation). - Option for single-vector delivery of multiple or complex antigens. | - RNA replicons are not able to tolerate many of the synthetic nucleotide modifications and sequence alterations. - Inclusion of unrelated proteins, which may increase unwanted immunogenicity. - Large replicon size (~10 kb), decreasing cell internalization efficiency. - Interaction between nsPs and host factors yet to be addressed. - Longer RNA length (more difficult production). - Potential elevated inflammation. | [8,13,29,86,93] |
| Name | Therapetic Modality | Protein Target | Administration Method | Delivery Vehicle | Disease | Sponsor Institution | ClinicalTrials.gov Identifer | Phase | Therapeutic Approach | References |
|---|---|---|---|---|---|---|---|---|---|---|
| MRT5005 | mRNA | CFTR | Inhalation | LNPs | Cystic fibrosis | Translate Bio | NCT03375047 | I/II | Protein Replacement | [94] |
| AZD8601 | mRNA | VEGF-A | Intracardiac injection | Naked mRNA | Heart failure | AstraZeneca | NCT03370887 | II | Cell reprogramming | [15] |
| CV7201 | mRNA | Rabies virus glycoprotein | I.D or I.M | RNActive, protamine | Rabies | CureVac | NCT02241135 | I | Immunotherapy | [95] |
| CV7202 | mRNA | Rabies virus glycoprotein | I.M | LNPs | Rabies | NCT03713086 | I | Immunotherapy | [13] | |
| CV9201 | mRNA | TAAs | I.D | RNActive, protamine | NSCLC | NCT00923312 | I/II | Immunotherapy | [96] | |
| CV9202 | mRNA | TAAs | I.D | RNActive, protamine | NSCLC | NCT03164772 | I/II | Immunotherapy | [8] | |
| CV9104 | mRNA | TAAs | I.D | RNActive, protamine | Prostate carcinoma | NCT02140138 | II | Immunotherapy | ||
| HARE-40 | mRNA | HPV antigen CD40 | I.D | Naked RNA | HPV-driven squamous cell carcinoma | BioNTech | NCT03418480 | I/II | Immunotherapy | |
| Lipo-MERIT | mRNA | TAAs: NYESO-1, MAGE-A3, tyrosinase, and TPTE | I.V | Lipo-MERIT, DOTMA(DOTAP)/ DOPE lipoplex | Advanced melanoma | NCT02410733 | I | Immunotherapy | ||
| IVAC | mRNA | 3 TAAs selected from a warehouse and p53 RNA; Neo-Ag based on NGS screening | I.V | Lipo-MERIT, DOTMA(DOTAP)/DOPE lipoplex | TNBC | BioNTech | NCT02316457 | I | Immunotherapy | [8] |
| RBL001/RBL002 | mRNA | TAAs | Ultrasound guided I.N | Naked mRNA | Melanoma | NCT01684241 | I | Immunotherapy | ||
| IVAC MUTANOME | mRNA | Neo-Ag | Ultrasound guided I.N | Naked mRNA | Melanoma | NCT02035956 | I | Immunotherapy | ||
| RO7198457 | mRNA | Neo-Ag | I.V | Naked mRNA | Melanoma; NSCLC; Bladder cancer | NCT03289962 | I | Immunotherapy | ||
| mRNA-1325 | mRNA | Zika virus antigen | I.D | LNPs | Zika virus | Moderna | NCT03014089 | I | Immunotherapy | |
| mRNA-1653 | mRNA | hMPV and hPIV type 3 vaccine | I.D | LNPs | hMPV and hPIV infection | NCT03392389 | I | Immunotherapy | ||
| VAL-506440 | mRNA | H10N8 antigen | I.D | LNPs | Influenza | Moderna | NCT03076385 | I | Immunotherapy | [97] |
| VAL-339851 | mRNA | H7 influenza antigen | I.D | LNPs | Influenza | NCT03345043 | I | Immunotherapy | ||
| mRNA-1647/1443 | mRNA | CMV glycoprotein H pentamer complex | I.D | LNPs | CMV infection | NCT03382405 | I | Immunotherapy | [8] | |
| mRNA-2416 | mRNA | Human OX40L | I.D | LNPs | Solid tumor malignancies or lymphoma | NCT03323398 | I | Immunotherapy | ||
| mRNA-4157 | mRNA | Neo-Ag | Intratumoral | LNPs | Solid tumor | NCT03313778 | I | Immunotherapy | ||
| mRNA-4650 | mRNA | Neo-Ag | I.M | Naked mRNA | Melanoma; Colon cancer; GI cancer; Genitourinary cancer; HCC | NCT03480152 | I/II | Immunotherapy | [98,99] | |
| mRNA-1388 | mRNA | VAL-181388 | I.M | LNPs | CHIKV | NCT03325075 | I | Immunotherapy | [12] | |
| mRNA-2752 | mRNA | OX40L, IL-23, and IL-36γ | Intratumoral | LNPs | Solid tumor or lymphoma | Moderna/AstraZeneca | NCT03739931 | I | Immunotherapy | [13] |
| iHIVARNA-01 | mRNA | Trimix (CD40L, CD70 and caTLR4 RNA—mRNA-transfected) | I.N | Naked mRNA | HIV infection | Hospital Clínic de Barcelona | NCT02413645 | I | Immunotherapy | [100] |
| mRNA | I.N | Naked mRNA | HIV infection | Erasmus Medical Center | NCT02888756 | II | Immunotherapy | [101] | ||
| - | mRNA | CT7, MAGE-A3, and WT1 mRNA-electroporated LCs | I.D | DC-loaded mRNA | Malignant melanoma | Memorial Sloan Kettering Cancer Center | NCT01995708 | I | Immunotherapy | [8] |
| - | mRNA | HIV-1 Gag- and Nef-transfected DCs | I.D | DC-loaded mRNA | HIV infection | Massachusetts General Hospital | NCT00833781 | I/II | Immunotherapy | [102] |
| - | mRNA | Neo-Ag | S.C | Naked mRNA | Solid tumor malignancies or lymphoma | Changhai Hospital Stemirna Therapeutics | NCT03468244 | N.A | Immunotherapy | [8] |
| - | mRNA | TAA for melanoma (Melan-A, MAGE-A1, MAGE-A3, survivin, GP100, and tyrosinase) | I.D | Naked mRNA | Melanoma | University Hospital Tuebingen | NCT00204516 | I/II | Immunotherapy | [12] |
| - | mRNA | TAA-transfected DC | I.D or I.N | DC-loaded mRNA | Malignant melanoma | Oslo University Hospital | NCT01278940 | I/II | Immunotherapy | [103] |
| - | mRNA | I.D | DC-loaded mRNA | Prostate cancer | NCT01278914 | I/II | Immunotherapy | [8] | ||
| AVX601 | Replicon | Alphavirus replicon vaccine expressing CMV genes | I.M or S.C | - | CMV | AlphaVax | NCT00439803 | I | Immunotherapy | [8] |
| AVX502 | Replicon | Alphavirus replicon vaccine expressing an influenza HA protein | I.M or S.C | - | Influenza | NCT00440362; NCT00706732 | I/II | Immunotherapy | ||
| AVX101 | Replicon | Alphavirus replicon, HIV-1 subtype C Gag vaccine | I.M or S.C | - | HIV infections | NCT00097838; NCT00063778 | I | Immunotherapy | [104] | |
| AVX701 | Replicon | Alphavirus replicon encoding the protein | I.M or S.C | - | Colon cancer; CRC; Breast cancer; Lung cancer; Pancreatic cancer | NCT01890213; NCT00529984 | I/II | Immunotherapy | [8] | |
| NY-ESO-1 | CRISPR-Cas9 | PD-1 and TCR | Ex vivo | Autologous T cells | Multiple myeloma; Synovial sarcoma; Melanoma | University of Pennsylvania | NCT03399448 | I | Gene Editing | [8] |
| CRISPR/TALEN-HPV E6/E7 | CRISPR/Cas9, TALEN | E6 and E7 | N.A | Plasmid DNA in gel | Cervical intraepithelial neoplasia | First Affiliated Hospital, Sun Yat-Sen University | NCT03057912 | I | Gene Editing | [105,106] |
| CTX001 | CRISPR-Cas9 | BCL11A | Ex vivo | Modified CD34+ hHSPCs | ß-thalassemia | Vertex Pharmaceuticals Incorporated | NCT03655678 | I/II | Gene Editing | [8] |
| - | CRISPR-Cas9 | PD-1 and TCR | Ex vivo | CAR-T cells | Mesothelin positive multiple solid tumors | Chinese PLA General Hospital | NCT03545815 | I | Gene Editing | |
| - | CRISPR-Cas9 | CD19 and CD20 | Ex vivo | Dual specificity CAR-T cells | ß cell leukemia and lymphoma | NCT03398967 | I/II | Gene Editing | ||
| UCART019 | CRISPR-Cas9 | CD19 | Ex vivo | CAR-T cells | ß cell leukemia and lymphoma | NCT03166878 | I/II | Gene Editing | [107] | |
| - | CRISPR-Cas9 | PD-1 | Ex vivo | Cytotoxic T lymphocytes | EBV-associated malignancies | Yang Yang | NCT03044743 | I/II | Gene Editing | [108,109] |
| SB-728mR-HSPC | ZFN mRNA | CCR5 | Ex vivo (mRNA) | CD34+ hHSPCs | HIV | City of Hope Medical Center | NCT02500849 | I | Gene Editing | [110] |
| SB-728mR-T | ZFN mRNA | CCR5 | Ex vivo (mRNA) | T cells | HIV | Sangamo Therapeutics | NCT02225665 | I/II | Gene Editing | [111] |
| Lipid Name | Role | Abbreviation | Molar Lipid Ratios (%) (Ionizable Cationic Lipid:Neutral Lipid:Cholesterol:PEG-ylated Lipid) |
|---|---|---|---|
| BNT162b2 vaccine | |||
| 4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate | ionizable cationic lipid | ALC-0315 | 46.3:9.4:42.7:1.6 |
| 1,2-Distearoyl-sn-glycero-3-phosphocholine | helper lipid | DSPC | |
| cholesterol | helper lipid | Chol | |
| 2-[(polyethylene glycol)-2000]-N,N-ditetradecylacetamide | PEG-lipid | ALC-0159 | |
| mRNA-1273 vaccine | |||
| heptadecan-9-yl 8-((2-hydroxyethyl)[6-oxo-6-(undecyloxy)hexyl]amino)octanoate | ionizable cationic lipid | SM-102 | 50:10:38.5:1.5 |
| 1,2-distearoyl-sn-glycero-3-phosphocholine | helper lipid | DSPC | |
| cholesterol | helper lipid | Chol | |
| 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 | PEG-lipid | PEG2000-DMG | |
| Name | Therapetic Modality | Protein Target | Administration Method | Delivery Vehicle | Developer | ClinicalTrials.gov Identifer, EU Clinical Trials Register or Chinese Clinical Trial Register | Phase |
|---|---|---|---|---|---|---|---|
| mRNA-1273 | mRNA | Spike Glycoprotein | Intramuscular | LNP | Moderna/NIAID | EUCTR2021-002327-38-NL | IV |
| BNT162b2 | mRNA | RBD/Spike Glycoprotein | Intramuscular | LNP | Pfizer/BioNTech + Fosun Pharma | NCT04760132 | IV |
| CVnCoV Vaccine | mRNA | Spike Glycoprotein | Intramuscular | LNP | CureVac AG | NCT04674189 | III |
| ARCT-021 | mRNA | Spike Glycoprotein | Intramuscular | LNP | Arcturus Therapeutics | NCT04668339 | II |
| LNP-nCoVsaRNA | mRNA | Spike Glycoprotein | Intramuscular | LNP | Imperial College London | ISRCTN17072692 | I |
| SARS-CoV-2 mRNA vaccine (ARCoV) | mRNA | RBD | Intramuscular | LNP | AMS/Walvax Biotechnology and Suzhou Abogen Biosciences | NCT04847102 | III |
| ChulaCov19 mRNA vaccine | mRNA | Spike Glycoprotein | Intramuscular | LNP | Chulalongkorn University | NCT04566276 | I |
| PTX-COVID19-B, mRNA vaccine | mRNA | Spike Glycoprotein | Intramuscular | LNP | Providence therapeutics | NCT04765436 | I |
| saRNA formulated in a NLC | mRNA | - | - | NLC | Infectious Disease Research Institute/Amyris, Inc. | - | Pre-Clinical |
| LNP-encapsulated mRNA encoding S | mRNA | Spike Glycoprotein | - | LNP | Max-Planck-Institute of Colloids and Interfaces | - | Pre-Clinical |
| Self-amplifying RNA | mRNA | - | - | - | Gennova | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | Selcuk University | - | Pre-Clinical |
| LNP-mRNA | mRNA | - | - | LNP | Translate Bio/Sanofi Pasteur | - | Pre-Clinical |
| LNP-mRNA | mRNA | - | - | LNP | CanSino Biologics/Precision NanoSystems | - | Pre-Clinical |
| LNP-encapsulated mRNA cocktail encoding VLP | mRNA | - | - | LNP | Fudan University/Shanghai JiaoTong University/RNACure Biopharma | - | Pre-Clinical |
| LNP-encapsulated mRNA encoding RBD | mRNA | RBD | - | LNP | Fudan University/Shanghai JiaoTong University/RNACure Biopharma | - | Pre-Clinical |
| Replicating Defective SARS-CoV-2 derived RNAs | mRNA | - | - | - | Centro Nacional Biotecnología (CNB-CSIC), Spain | - | Pre-Clinical |
| LNP-encapsulated mRNA | mRNA | - | - | LNP | University of Tokyo/Daiichi-Sankyo | - | Pre-Clinical |
| Liposome-encapsulated mRNA | mRNA | - | - | LNP | BIOCAD | - | Pre-Clinical |
| Several mRNA candidates | mRNA | - | - | - | RNAimmune, Inc. | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | FBRI SRC VB VECTOR, Rospotrebnadzor, Koltsovo | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | China CDC/Tongji University/Stermina | - | Pre-Clinical |
| mRNA in an intranasal delivery system | mRNA | - | Intranasal | - | eTheRNA | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | Greenlight Biosciences | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | IDIBAPS-Hospital Clinic, Spain | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | Providence Therapeutics | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | Cell Tech Pharmed | - | Pre-Clinical |
| mRNA | mRNA | - | - | - | ReNAP Co. | - | Pre-Clinical |
| D614G variant LNP-encapsulated mRNA | mRNA | - | - | LNP | Globe Biotech Ltd. | - | Pre-Clinical |
| Encapsulated mRNA | mRNA | - | - | - | CEA | - | Pre-Clinical |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baptista, B.; Carapito, R.; Laroui, N.; Pichon, C.; Sousa, F. mRNA, a Revolution in Biomedicine. Pharmaceutics 2021, 13, 2090. https://doi.org/10.3390/pharmaceutics13122090
Baptista B, Carapito R, Laroui N, Pichon C, Sousa F. mRNA, a Revolution in Biomedicine. Pharmaceutics. 2021; 13(12):2090. https://doi.org/10.3390/pharmaceutics13122090
Chicago/Turabian StyleBaptista, Bruno, Rita Carapito, Nabila Laroui, Chantal Pichon, and Fani Sousa. 2021. "mRNA, a Revolution in Biomedicine" Pharmaceutics 13, no. 12: 2090. https://doi.org/10.3390/pharmaceutics13122090
APA StyleBaptista, B., Carapito, R., Laroui, N., Pichon, C., & Sousa, F. (2021). mRNA, a Revolution in Biomedicine. Pharmaceutics, 13(12), 2090. https://doi.org/10.3390/pharmaceutics13122090