Design and Validation of a Process Based on Cationic Niosomes for Gene Delivery into Novel Urine-Derived Mesenchymal Stem Cells
Abstract
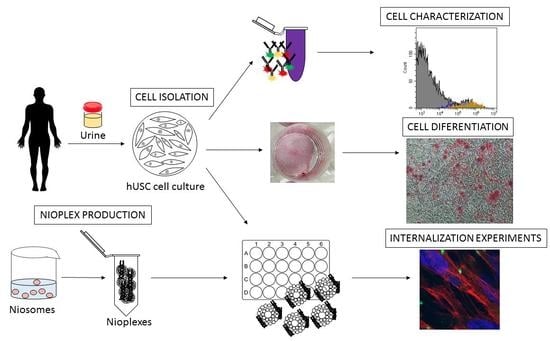
:1. Introduction
2. Materials and Methods
2.1. Cell Culture Isolation
2.2. Cell Characterization
- •
- Surface markers specific for mesenchymal stem cells: antiCD73-AlexaFluor488, antiCD90-APC, and antiCD105-PE;
- •
- Surface markers that are absent in mesenchymal stem cells: antiCD14-FITC, antiCD34-APC, and antiCD45-PE.
2.3. Plasmid Propagation and Elaboration of Nioplexes
2.4. Physicochemical Characterization of Cationic Niosomes/Nioplexes
2.5. Cell Culture and In Vitro Transfection Assays
2.6. Cell Uptake and Intracellular Distribution of Nioplexes
2.7. Intracellular Trafficking Studies
3. Results and Discussion
3.1. Cell Culture Isolation
3.2. Cell Characterization
3.3. Physicochemical Characterization of Cationic Niosomes/Nioplexes
3.4. Cell Viability and Transfection Efficiency of Nioplexes
3.5. Cellular Uptake Studies
3.6. Trafficking of the Nioplex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Verma, I.M.; Weitzman, M.D. Gene therapy: Twenty-first century medicine. Annu. Rev. Biochem. 2005, 74, 711–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Deriglasova, U.F.; Kulagina, N.N.; Panasuk, A.F.; Rudakowa, S.F.; Luriá, E.A.; Ruadkow, I.A. Precursors for fibroblasts in different populations of hematopoietic cells as detected by the in vitro colony assay method. Exp. Hematol. 1974, 2, 83–92. [Google Scholar] [PubMed]
- Jeon, B.-G.; Jang, S.-J.; Park, J.-S.; Subbarao, R.B.; Jeong, G.-J.; Park, B.-W.; Rho, G.-J. Differentiation potential of mesenchymal stem cells isolated from human dental tissues into non-mesodermal lineage. Anim. Cells Syst. 2015, 19, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Dominici, M.; Blanc, K.L.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Horwitz, E.M.; Blanc, K.L.; Dominici, M.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Deans, R.J.; Krause, D.S.; Keating, A. International Society for Cellular Therapy Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy 2005, 7, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Attia, N.; Mashal, M.; Grijalvo, S.; Eritja, R.; Zárate, J.; Puras, G.; Pedraz, J.L. Stem cell-based gene delivery mediated by cationic niosomes for bone regeneration. Nanomedicine 2018, 14, 521–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Li, L.; Xing, F.; Peng, J.; Peng, K.; Wang, Y.; Xiang, Z. Human urine-derived stem cells: Potential for cell-based therapy of cartilage defects. Stem Cells Int. 2018, 2018, 4686259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, X.; Wang, M.; Chen, F.; Zhou, J. Urine-derived stem cells: The present and the future. Stem Cells Int. 2017, 2017, 4378947. [Google Scholar] [CrossRef] [Green Version]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.; Futrell, J.W.; Katz, A.J.; Benhaim, P.; Lorenz, H.P.; Hedrick, M.H. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001, 7, 211–228. [Google Scholar] [CrossRef] [Green Version]
- Sarugaser, R.; Lickorish, D.; Baksh, D.; Hosseini, M.M.; Davies, J.E. Human umbilical cord perivascular (HUCPV) cells: A source of mesenchymal progenitors. Stem Cells 2005, 23, 220–229. [Google Scholar] [CrossRef]
- Guan, J.-J.; Niu, X.; Gong, F.-X.; Hu, B.; Guo, S.-C.; Lou, Y.-L.; Zhang, C.-Q.; Deng, Z.-F.; Wang, Y. Biological characteristics of human-urine-derived stem cells: Potential for cell-based therapy in neurology. Tissue Eng. Part A 2014, 20, 1794–1806. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; McNeill, E.; Tian, H.; Soker, S.; Andersson, K.-E.; Yoo, J.J.; Atala, A. Urine derived cells are a potential source for urological tissue reconstruction. J. Urol. 2008, 180, 2226–2233. [Google Scholar] [CrossRef] [PubMed]
- Bento, G.; Shafigullina, A.K.; Rizvanov, A.A.; Sardão, V.A.; Macedo, M.P.; Oliveira, P.J. Urine-derived stem cells: Applications in regenerative and predictive medicine. Cells 2020, 9, 573. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, Y.; Bharadwaj, S.; Atala, A.; Zhang, Y. Human urine-derived stem cells seeded in a modified 3D porous small intestinal submucosa scaffold for urethral tissue engineering. Biomaterials 2011, 32, 1317–1326. [Google Scholar] [CrossRef]
- Qin, H.; Zhu, C.; An, Z.; Jiang, Y.; Zhao, Y.; Wang, J.; Liu, X.; Hui, B.; Zhang, X.; Wang, Y. Silver nanoparticles promote osteogenic differentiation of human urine-derived stem cells at noncytotoxic concentrations. Int. J. Nanomed. 2014, 9, 2469–2478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zheng, Y.; Li, S.; Xue, H.; Schmitt, K.; Hergenroeder, G.W.; Wu, J.; Zhang, Y.; Kim, D.H.; Cao, Q. Human neural progenitors derived from integration-free iPSCs for SCI therapy. Stem Cell Res. 2017, 19, 55–64. [Google Scholar] [CrossRef]
- Guan, J.; Zhang, J.; Zhu, Z.; Niu, X.; Guo, S.; Wang, Y.; Zhang, C. Bone morphogenetic protein 2 gene transduction enhances the osteogenic potential of human urine-derived stem cells. Stem Cell Res. Ther. 2015, 6, 5. [Google Scholar] [CrossRef] [Green Version]
- Reiser, J.; Zhang, X.-Y.; Hemenway, C.S.; Mondal, D.; Pradhan, L.; Russa, V.F.L. Potential of mesenchymal stem cells in gene therapy approaches for inherited and acquired diseases. Expert Opin. Biol. Ther. 2005, 5, 1571–1584. [Google Scholar] [CrossRef]
- Bharadwaj, S.; Liu, G.; Shi, Y.; Wu, R.; Yang, B.; He, T.; Fan, Y.; Lu, X.; Zhou, X.; Liu, H.; et al. Multipotential differentiation of human urine-derived stem cells: Potential for therapeutic applications in urology. Stem Cells 2013, 31, 1840–1856. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.; Liu, G.; Shi, Y.; Bharadwaj, S.; Leng, X.; Zhou, X.; Liu, H.; Atala, A.; Zhang, Y. Self-renewal and differentiation capacity of urine-derived stem cells after urine preservation for 24 h. PLoS ONE 2013, 8, e53980. [Google Scholar] [CrossRef] [Green Version]
- Baum, C.; Kustikova, O.; Modlich, U.; Li, Z.; Fehse, B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum. Gene Ther. 2006, 17, 253–263. [Google Scholar] [CrossRef]
- Bessis, N.; GarciaCozar, F.J.; Boissier, M.-C. Immune responses to gene therapy vectors: Influence on vector function and effector mechanisms. Gene Ther. 2004, 11, S10–S17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Bouard, D.; Alazard-Dany, D.; Cosset, F.-L. Viral vectors: From virology to transgene expression. Br. J. Pharmacol. 2009, 157, 153–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef]
- Putnam, D. Polymers for gene delivery across length scales. Nat. Mater. 2006, 5, 439–451. [Google Scholar] [CrossRef]
- Pezzoli, D.; Kajaste-Rudnitski, A.; Chiesa, R.; Candiani, G. Lipid-Based Nanoparticles as Nonviral Gene Delivery Vectors. In Nanomaterial Interfaces in Biology; Humana Press: Totowa, NJ, USA, 2013; pp. 269–279. [Google Scholar]
- Rajera, R.; Nagpal, K.; Singh, S.K.; Mishra, D.N. Niosomes: A controlled and novel drug delivery system. Biol. Pharm. Bull. 2011, 34, 945–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda, E.; Agirre, M.; Villate-Beitia, I.; Mashal, M.; Puras, G.; Zarate, J.; Pedraz, J.L. Elaboration and physicochemical characterization of niosome-based nioplexes for gene delivery purposes. Methods Mol. Biol. 2016, 1445, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Puras, G.; Mashal, M.; Zárate, J.; Agirre, M.; Ojeda, E.; Grijalvo, S.; Eritja, R.; Diaz-Tahoces, A.; Martínez Navarrete, G.; Avilés-Trigueros, M.; et al. A novel cationic niosome formulation for gene delivery to the retina. J. Control. Release 2014, 174, 27–36. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, S.; Jiang, H.; Zhao, B.; Lv, H. Lipoplex morphologies and their influences on transfection efficiency in gene delivery. J. Control. Release 2007, 123, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yang, J.; Huang, L.; Liu, D. Effect of non-ionic surfactants on the formation of DNA/emulsion complexes and emulsion-mediated gene transfer. Pharm. Res. 1996, 13, 1642–1646. [Google Scholar] [CrossRef]
- Moghassemi, S.; Hadjizadeh, A. Nano-niosomes as nanoscale drug delivery systems: An illustrated review. J. Control. Release 2014, 185, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Kim, T.W.; Kwon, M.; Kwon, I.C.; Jeong, S.Y. Oil components modulate physical characteristics and function of the natural oil emulsions as drug or gene delivery system. J. Control. Release 2001, 71, 339–350. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Rao, S.-S.; Ren, L.; Hu, X.-K.; Tan, Y.-J.; Hu, Y.; Luo, J.; Liu, Y.-W.; Yin, H.; Huang, J.; et al. Exosomal DMBT1 from human urine-derived stem cells facilitates diabetic wound repair by promoting angiogenesis. Theranostics 2018, 8, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Hulspas, R.; O’Gorman, M.R.G.; Wood, B.L.; Gratama, J.W.; Sutherland, D.R. Considerations for the control of background fluorescence in clinical flow cytometry. Cytom. B. Clin. Cytom. 2009, 76, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Gallego, I.; Villate-Beitia, I.; Martínez-Navarrete, G.; Menéndez, M.; López-Méndez, T.; Soto-Sánchez, C.; Zárate, J.; Puras, G.; Fernández, E.; Pedraz, J.L. Non-viral vectors based on cationic niosomes and minicircle DNA technology enhance gene delivery efficiency for biomedical applications in retinal disorders. Nanomedicine 2019, 17, 308–318. [Google Scholar] [CrossRef]
- Mashal, M.; Attia, N.; Puras, G.; Martínez-Navarrete, G.; Fernández, E.; Pedraz, J.L. Retinal gene delivery enhancement by lycopene incorporation into cationic niosomes based on DOTMA and polysorbate. J. Control. Release 2017, 254, 55–64. [Google Scholar] [CrossRef]
- Ojeda, E.; Puras, G.; Agirre, M.; Zarate, J.; Grijalvo, S.; Eritja, R.; DiGiacomo, L.; Caracciolo, G.; Pedraz, J.-L. The role of helper lipids in the intracellular disposition and transfection efficiency of niosome formulations for gene delivery to retinal pigment epithelial cells. Int. J. Pharm. 2016, 503, 115–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villate-Beitia, I.; Gallego, I.; Martínez-Navarrete, G.; Zárate, J.; López-Méndez, T.; Soto-Sánchez, C.; Santos-Vizcaíno, E.; Puras, G.; Fernández, E.; Pedraz, J.L. Polysorbate 20 non-ionic surfactant enhances retinal gene delivery efficiency of cationic niosomes after intravitreal and subretinal administration. Int. J. Pharm. 2018, 550, 388–397. [Google Scholar] [CrossRef]
- Van Steensel, B.; Van Binnendijk, E.P.; Hornsby, C.D.; Van der Voort, H.T.; Krozowski, Z.S.; Kloet, E.R.D.; Van Driel, R. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 1996, 109, 787–792. [Google Scholar] [CrossRef]
- Andar, A.U.; Hood, R.R.; Vreeland, W.N.; DeVoe, D.L.; Swaan, P.W. Microfluidic preparation of liposomes to determine particle size influence on cellular uptake mechanisms. Pharm. Res. 2014, 31, 401–413. [Google Scholar] [CrossRef]
- Hosseinkhani, H.; Tabata, Y. Self assembly of DNA nanoparticles with polycations for the delivery of genetic materials into cells. J. Nanosci. Nanotechnol. 2006, 6, 2320–2328. [Google Scholar] [CrossRef] [PubMed]
- Rezvani, A.Z.; Rahimizadeh, M.; Eshghi, H.; Dehshahri, A.; Ramezani, M. The effect of cationic charge density change on transfection efficiency of polyethylenimine. Iran J. Basic Med. Sci. 2013, 16, 150–156. [Google Scholar]
- Kumar, A.; Dixit, C.K. Methods for characterization of nanoparticles. In Advances in Nanomedicine for the Delivery of Therapeutic Nucleic Acids; Elsevier: Amsterdam, The Netherlands, 2017; pp. 43–58. [Google Scholar]
- Nafee, N.; Taetz, S.; Schneider, M.; Schaefer, U.F.; Lehr, C.-M. Chitosan-coated PLGA nanoparticles for DNA/RNA delivery: Effect of the formulation parameters on complexation and transfection of antisense oligonucleotides. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Naseri, N.; Valizadeh, H.; Zakeri-Milani, P. Solid lipid nanoparticles and nanostructured lipid carriers: Structure, preparation and application. Adv. Pharm. Bull. 2015, 5, 305–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radler, J.O. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science 1997, 275, 810–814. [Google Scholar] [CrossRef] [Green Version]
- Pawlowska, D.; Janich, C.; Langner, A.; Dobner, B.; Wölk, C.; Brezesinski, G. The impact of alkyl-chain purity on lipid-based nucleic acid delivery systems—Is the utilization of lipid components with technical grade justified? Chemphyschem 2019, 20, 2110–2121. [Google Scholar] [CrossRef]
- Eygeris, Y.; Patel, S.; Jozic, A.; Sahay, G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020, 20, 4543–4549. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, T.G.D.; Pellenz, F.M.; Laureano, A.; Silla, L.M.R.D.; Giugliani, R.; Baldo, G.; Matte, U. A simple protocol for transfecting human mesenchymal stem cells. Biotechnol. Lett. 2018, 40, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.D.; Andrade, P.Z.; Abecasis, M.M.; Gimble, J.M.; Chase, L.G.; Campbell, A.M.; Boucher, S.; Vemuri, M.C.; Silva, C.L.D.; Cabral, J.M.S. Toward a clinical-grade expansion of mesenchymal stem cells from human sources: A microcarrier-based culture system under xeno-free conditions. Tissue Eng. Part C Methods 2011, 17, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, X.; Li, Z.; Lendlein, A.; Ma, N. Genetic engineering of mesenchymal stem cells by non-viral gene delivery. Clin. Hemorheol. Microcirc. 2014, 58, 19–48. [Google Scholar] [CrossRef] [Green Version]
- Villate-Beitia, I.; Puras, G.; Soto-Sánchez, C.; Agirre, M.; Ojeda, E.; Zarate, J.; Fernández, E.; Pedraz, J.L. Non-viral vectors based on magnetoplexes, lipoplexes and polyplexes for VEGF gene delivery into central nervous system cells. Int. J. Pharm. 2017, 521, 130–140. [Google Scholar] [CrossRef]
- Deshpande, P.P.; Biswas, S.; Torchilin, V.P. Current trends in the use of liposomes for tumor targeting. Nanomedicine 2013, 8, 1509–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda, E.; Puras, G.; Agirre, M.; Zarate, J.; Grijalvo, S.; Eritja, R.; Martinez-Navarrete, G.; Soto-Sánchez, C.; Diaz-Tahoces, A.; Aviles-Trigueros, M.; et al. The influence of the polar head-group of synthetic cationic lipids on the transfection efficiency mediated by niosomes in rat retina and brain. Biomaterials 2016, 77, 267–279. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Sawyer, G.J.; Dong, X.; Qiu, Y.; Collins, L.; Fabre, J.W. The in vivo use of chloroquine to promote non-viral gene delivery to the liver via the portal vein and bile duct. J. Gene Med. 2003, 5, 209–218. [Google Scholar] [CrossRef]
- Rejman, J.; Oberle, V.; Zuhorn, I.S.; Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef]
- Xiang, S.; Tong, H.; Shi, Q.; Fernandes, J.C.; Jin, T.; Dai, K.; Zhang, X. Uptake mechanisms of non-viral gene delivery. J. Control. Release 2012, 158, 371–378. [Google Scholar] [CrossRef]
- Wasungu, L.; Hoekstra, D. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Control. Release 2006, 116, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Nichols, B. Caveosomes and endocytosis of lipid rafts. J. Cell Sci. 2003, 116, 4707–4714. [Google Scholar] [CrossRef] [Green Version]
- Xiang, S.; Zhang, X. Cellular Uptake Mechanism of Non-Viral Gene Delivery and Means for Improving Transfection Efficiency. In Gene Therapy—Tools and Potential Applications; InTech: London, UK, 2013. [Google Scholar]
- Sanz, V.; Coley, H.M.; Silva, S.R.P.; McFadden, J. Protamine and chloroquine enhance gene delivery and expression mediated by RNA-wrapped single walled carbon nanotubes. J. Nanosci. Nanotechnol. 2012, 12, 1739–1747. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zeidan, R.; Mishra, S.; Liu, A.; Pun, S.H.; Kulkarni, R.P.; Jensen, G.S.; Bellocq, N.C.; Davis, M.E. Structure-function correlation of chloroquine and analogues as transgene expression enhancers in nonviral gene delivery. J. Med. Chem. 2006, 49, 6522–6531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashal, M.; Attia, N.; Soto-Sánchez, C.; Martínez-Navarrete, G.; Fernández, E.; Puras, G.; Pedraz, J.L. Non-viral vectors based on cationic niosomes as efficient gene delivery vehicles to central nervous system cells into the brain. Int. J. Pharm. 2018, 552, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; He, H.; Liu, H.; Thayumanavan, S. Cellular uptake evaluation of amphiphilic polymer assemblies: Importance of interplay between pharmacological and genetic approaches. Biomacromolecules 2019, 20, 4407–4418. [Google Scholar] [CrossRef]
- Ho, L.W.C.; Yung, W.-Y.; Sy, K.H.S.; Li, H.Y.; Choi, C.K.K.; Leung, K.C.-F.; Lee, T.W.Y.; Choi, C.H.J. Effect of alkylation on the cellular uptake of polyethylene glycol-coated gold nanoparticles. ACS Nano 2017, 11, 6085–6101. [Google Scholar] [CrossRef] [PubMed]





| Cell Type | CD73-AlexaFluor488 | CD90-APC | CD105-PE | CD14-FITC | CD34-APC | CD45-PE |
|---|---|---|---|---|---|---|
| Commercial mesenchymal stem cells | 63.4% | 81.7% | 70.1% | 0.8% | 1.3% | 20.5% |
| hUSCs | 78.6% | 86.4% | 66.1% | 6.8% | 13.4% | 52.4% |
| HEK293 | 36.5% | 0.9% | 3.5% | 2.8% | 0.8% | 11.3% |
| Formulation | Size (nm) | Zeta Potential (mV) | PdI |
|---|---|---|---|
| GPxT-CQ | 109.8 ± 1.01 | 33.4 ± 5.7 | 0.13 ± 0.01 |
| GPxT-CQ/DNA (5:1) | 162.3 ± 2.6 | 21.2 ± 2.4 | 0.31 ± 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vado, Y.; Puras, G.; Rosique, M.; Martin, C.; Pedraz, J.L.; Jebari-Benslaiman, S.; de Pancorbo, M.M.; Zarate, J.; Perez de Nanclares, G. Design and Validation of a Process Based on Cationic Niosomes for Gene Delivery into Novel Urine-Derived Mesenchymal Stem Cells. Pharmaceutics 2021, 13, 696. https://doi.org/10.3390/pharmaceutics13050696
Vado Y, Puras G, Rosique M, Martin C, Pedraz JL, Jebari-Benslaiman S, de Pancorbo MM, Zarate J, Perez de Nanclares G. Design and Validation of a Process Based on Cationic Niosomes for Gene Delivery into Novel Urine-Derived Mesenchymal Stem Cells. Pharmaceutics. 2021; 13(5):696. https://doi.org/10.3390/pharmaceutics13050696
Chicago/Turabian StyleVado, Yerai, Gustavo Puras, Melania Rosique, Cesar Martin, Jose Luis Pedraz, Shifa Jebari-Benslaiman, Marian M. de Pancorbo, Jon Zarate, and Guiomar Perez de Nanclares. 2021. "Design and Validation of a Process Based on Cationic Niosomes for Gene Delivery into Novel Urine-Derived Mesenchymal Stem Cells" Pharmaceutics 13, no. 5: 696. https://doi.org/10.3390/pharmaceutics13050696
APA StyleVado, Y., Puras, G., Rosique, M., Martin, C., Pedraz, J. L., Jebari-Benslaiman, S., de Pancorbo, M. M., Zarate, J., & Perez de Nanclares, G. (2021). Design and Validation of a Process Based on Cationic Niosomes for Gene Delivery into Novel Urine-Derived Mesenchymal Stem Cells. Pharmaceutics, 13(5), 696. https://doi.org/10.3390/pharmaceutics13050696