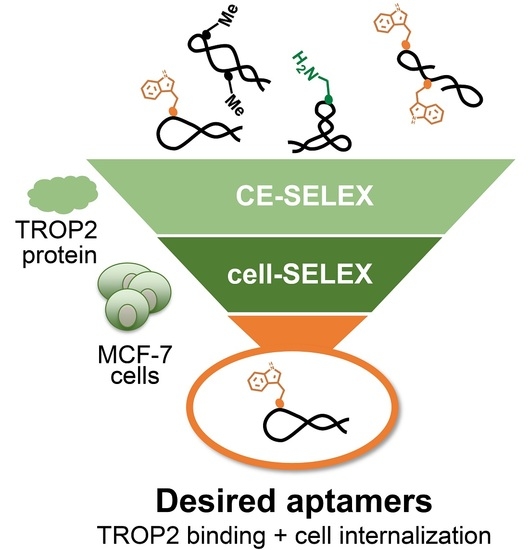
Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagents and Cell Lines
2.2. Preparation of the Initial Artificial Nucleic Acid Aptamer Libraries
2.3. Pre-Screening of Aptamer Libraries
2.4. CE-SELEX Process
2.5. Cell-SELEX Process
2.6. Sequencing and Bioinformatic Analysis of Aptamer Libraries
2.7. Preparation of Aptamers
2.8. Binding Affinity Analysis with CE
2.9. Cell Internalization Assays Using the qPCR Method
2.10. Statistical Analysis
3. Results
3.1. Pre-Screening of Artificial Nucleic Acid Libraries Using CE
3.2. CE-SELEX for Anti-TROP2 Aptamer Selection
3.3. Binding Affinity and Specificity of Aptamers Selected with CE-SELEX
3.4. Cell-SELEX for the Enrichment of Internalizing Aptamers against MCF-7 Cells
3.5. Binding and Specificity of Aptamers Selected with Cell-SELEX
3.6. Evaluation of Capacity to Undergo Cell Internalization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Kiel, J.L. Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. Biotechniques 2002, 32, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, S.D.; Bowser, M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 2004, 126, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Mendonsa, S.D.; Bowser, M.T. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal. Chem. 2004, 76, 5387–5392. [Google Scholar] [CrossRef]
- Kasahara, Y.; Irisawa, Y.; Fujita, H.; Yahara, A.; Ozaki, H.; Obika, S.; Kuwahara, M. Capillary electrophoresis–systematic evolution of ligands by exponential enrichment selection of base- and sugar-modified DNA aptamers: Target binding dominated by 2′-O,4′-C-Methylene-bridged/locked nucleic acid primer. Anal. Chem. 2013, 85, 4961–4967. [Google Scholar] [CrossRef] [PubMed]
- Wakui, K.; Yoshitomi, T.; Yamaguchi, A.; Tsuchida, M.; Saito, S.; Shibukawa, M.; Furusho, H.; Yoshimoto, K. Rapidly neutralizable and highly anticoagulant thrombin-binding DNA aptamer discovered by MACE SELEX. Mol. Ther. Nucleic Acids 2019, 16, 348–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdés, J.J.; Chambers, J.P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2005, 384, 191–198. [Google Scholar] [CrossRef]
- Lou, X.; Qian, J.; Xiao, Y.; Viel, L.; Gerdon, A.E.; Lagally, E.T.; Atzberger, P.; Tarasow, T.M.; Heeger, A.J.; Soh, H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 2009, 106, 2989–2994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, G.; Ahmed, M.-S.L.; Dolf, A.; Endl, E.; Knolle, P.A.; Famulok, M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010, 5, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, X.; Fu, X.; Li, Z.; Huang, Y.; Liang, C. Advances in aptamer-based biomarker discovery. Front. Cell Dev. Biol. 2021, 9, 571. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P. Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens. Mol. 2017, 22, 215. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Sullenger, B.A.; White, R.R. Further characterization of the target of a potential aptamer biomarker for pancreatic cancer: Cyclophilin B and its posttranslational modifications. Nucleic Acid Ther. 2013, 23, 435–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, D.A.; Chen, H.; Hicke, B.J.; Swiderek, K.M.; Gold, L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 2003, 100, 15416–15421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Yu, Y.; Jiang, F.; Zhou, J.; Li, Y.; Liang, C.; Dang, L.; Lu, A.; Zhang, G. Development of Cell-SELEX technology and its application in cancer diagnosis and therapy. Int. J. Mol. Sci. 2016, 17, 2079. [Google Scholar] [CrossRef] [Green Version]
- Gray, B.P.; Kelly, L.; Ahrens, D.P.; Barry, A.P.; Kratschmer, C.; Levy, M.; Sullenger, B.A. Tunable cytotoxic aptamer–drug conjugates for the treatment of prostate cancer. Proc. Natl. Acad. Sci. USA 2018, 115, 4761–4766. [Google Scholar] [CrossRef] [Green Version]
- Kratschmer, C.; Levy, M. Targeted delivery of auristatin-modified toxins to pancreatic cancer using aptamers. Mol. Ther. Nucleic Acids 2018, 10, 227–236. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.; Sun, N.; Liu, M.; Wang, J.; Pei, R. Building a chimera of aptamer–antisense oligonucleotide for silencing galectin-1 gene. RSC Adv. 2016, 6, 112445–112450. [Google Scholar] [CrossRef]
- Soldevilla, M.M.; De Caso, D.M.-C.; Menon, A.P.; Pastor, F. Aptamer-iRNAs as therapeutics for cancer treatment. Pharmaceuticals 2018, 11, 108. [Google Scholar] [CrossRef] [Green Version]
- Xuan, W.; Peng, Y.; Deng, Z.; Peng, T.; Kuai, H.; Li, Y.; He, J.; Jin, C.; Liu, Y.; Wang, R.; et al. A basic insight into aptamer-drug conjugates (ApDCs). Biomaterials 2018, 182, 216–226. [Google Scholar] [CrossRef]
- Mercier, M.-C.; Dontenwill, M.; Choulier, L. Selection of nucleic acid aptamers targeting tumor cell-surface protein biomarkers. Cancers 2017, 9, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O.; Giangrande, P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. [Google Scholar] [CrossRef] [Green Version]
- Parekh, P.; Kamble, S.; Zhao, N.; Zeng, Z.; Portier, B.P.; Zu, Y. Immunotherapy of CD30-expressing lymphoma using a highly stable ssDNA aptamer. Biomaterials 2013, 34, 8909–8917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iaboni, M.; Fontanella, R.; Rienzo, A.; Capuozzo, M.; Nuzzo, S.; Santamaria, G.; Catuogno, S.; Condorelli, G.; de Franciscis, V.; Esposito, C.L. Targeting Insulin Receptor with a Novel Internalizing Aptamer. Mol. Ther. Nucleic Acids 2016, 5, e365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef]
- Gupta, S.; Drolet, D.W.; Wolk, S.K.; Waugh, S.M.; Rohloff, J.C.; Carter, J.D.; Mayfield, W.S.; Otis, M.R.; Fowler, C.R.; Suzuki, T.; et al. Pharmacokinetic properties of DNA aptamers with base modifications. Nucleic Acid Ther. 2017, 27, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Zheng, C.; Zhou, S.-F.; Qiao, S.; Tran, P.H.-L.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; et al. Superior performance of aptamer in tumor penetration over antibody: implication of aptamer-based theranostics in solid tumors. Theranostics 2015, 5, 1083–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeiffer, F.; Rosenthal, M.; Siegl, J.; Ewers, J.; Mayer, G. Customised nucleic acid libraries for enhanced aptamer selection and performance. Curr. Opin. Biotechnol. 2017, 48, 111–118. [Google Scholar] [CrossRef]
- Imaizumi, Y.; Kasahara, Y.; Fujita, H.; Kitadume, S.; Ozaki, H.; Endoh, T.; Kuwahara, M.; Sugimoto, N. Efficacy of base-modification on target binding of small molecule DNA aptamers. J. Am. Chem. Soc. 2013, 135, 9412–9419. [Google Scholar] [CrossRef]
- Tanaka, K.; Kasahara, Y.; Miyamoto, Y.; Takumi, O.; Kasai, T.; Onodera, K.; Kuwahara, M.; Oka, M.; Yoneda, Y.; Obika, S. Development of oligonucleotide-based antagonists of Ebola virus protein 24 inhibiting its interaction with karyopherin alpha 1. Org. Biomol. Chem. 2018, 16, 4456–4463. [Google Scholar] [CrossRef]
- Hoshino, H.; Kasahara, Y.; Kuwahara, M.; Obika, S. DNA Polymerase Variants with High Processivity and Accuracy for Encoding and Decoding Locked Nucleic Acid Sequences. J. Am. Chem. Soc. 2020, 142, 21530–21537. [Google Scholar] [CrossRef]
- Gawande, B.N.; Rohloff, J.C.; Carter, J.D.; von Carlowitz, I.; Zhang, C.; Schneider, D.J.; Janjic, N. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. USA 2017, 114, 2898–2903. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Huang, J.-F.; Qiu, J.-R.; Zhang, H.-L.; Tang, X.-J.; Li, H.; Wang, C.-J.; Wang, Z.-C.; Feng, Z.-Q.; Zhu, J. Significantly upregulated TACSTD2 and Cyclin D1 correlate with poor prognosis of invasive ductal breast cancer. Exp. Mol. Pathol. 2013, 94, 73–78. [Google Scholar] [CrossRef]
- Shih, L.B.; Xuan, H.; Aninipot, R.; Stein, R.; Goldenberg, D.M. In vitro and in vivo reactivity of an internalizing antibody, RS7, with human breast cancer. Cancer Res. 1995, 55, 5857s–5863s. [Google Scholar] [PubMed]
- Goldenberg, D.M.; Cardillo, T.M.; Govindan, S.V.; Rossi, E.A.; Sharkey, R.M. Trop-2 is a novel target for solid cancer therapy with sacituzumab govitecan (IMMU-132), an antibody-drug conjugate (ADC)*. Oncotarget 2015, 6, 22496–22512. [Google Scholar] [CrossRef] [Green Version]
- Thiel, W.H.; Thiel, K.W.; Flenker, K.S.; Bair, T.; Dupuy, A.J.; McNamara, J.O.; Miller, F.J.; Giangrande, P.H. Cell-internalization SELEX: Method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol. Biol. 2014, 1218, 187–199. [Google Scholar] [CrossRef] [Green Version]
- Hoinka, J.; Przytycka, T. AptaPLEX—A dedicated, multithreaded demultiplexer for HT-SELEX data. Methods 2016, 106, 82–85. [Google Scholar] [CrossRef]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, M.; Mao, A.; Zeng, J.; Liu, Y.; Wang, X.; Ma, J.; Tian, Y.; Ma, N.; Yang, N.; et al. Target replacement strategy for selection of DNA aptamers against the Fc region of mouse IgG. Genet. Mol. Res. 2013, 12, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.; Kuznetsov, A. Inside the black box: What makes SELEX better? Molecules 2019, 24, 3598. [Google Scholar] [CrossRef] [Green Version]
- Hoinka, J.; Backofen, R.; Przytycka, T.M. AptaSUITE: A full-featured bioinformatics framework for the comprehensive analysis of aptamers from HT-SELEX experiments. Mol. Ther. Nucleic Acids 2018, 11, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Ambrogi, F.; Fornili, M.; Boracchi, P.; Trerotola, M.; Relli, V.; Simeone, P.; La Sorda, R.; Lattanzio, R.; Querzoli, P.; Pedriali, M.; et al. Trop-2 Is a Determinant of Breast Cancer Survival. PLoS ONE 2014, 9, e96993. [Google Scholar] [CrossRef] [Green Version]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jiménez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D.; et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef] [Green Version]
- Futami, K.; Kimoto, M.; Lim, Y.W.S.; Hirao, I. Genetic alphabet expansion provides versatile specificities and activities of unnatural-base DNA aptamers targeting cancer cells. Mol. Ther. Nucleic Acids 2019, 14, 158–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, D.R.; Gelinas, A.D.; Zhang, C.; Rohloff, J.C.; Carter, J.D.; O’Connell, D.; Waugh, S.M.; Wolk, S.K.; Mayfield, W.S.; Burgin, A.B.; et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. USA 2012, 109, 19971–19976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolot, R.; Lam, C.H.; Sierant, M.; Zhao, Q.; Liu, F.-W.; Nawrot, B.; Egli, M.; Yang, X. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018, 46, 4819–4830. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Okuda, T.; Kasahara, Y.; Obika, S. Base-modified aptamers obtained by cell-internalization SELEX facilitate cellular uptake of an antisense oligonucleotide. Mol. Ther. Nucleic Acids 2021, 23, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Uemachi, H.; Kasahara, Y.; Tanaka, K.; Okuda, T.; Yoneda, Y.; Obika, S. Discovery of cell-internalizing artificial nucleic acid aptamers for lung fibroblasts and targeted drug delivery. Bioorganic Chem. 2020, 105, 104321. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Shi, Y.-S.; Wu, X.-D.; Liang, H.-Y.; Gao, Y.-B.; Li, S.-J.; Zhang, X.; Wang, F.; Gao, T.-M. DNA aptamers that target human glioblastoma multiforme cells overexpressing epidermal growth factor receptor variant III in vitro. Acta Pharmacol. Sin. 2013, 34, 1491–1498. [Google Scholar] [CrossRef] [Green Version]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef] [PubMed]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Al Bawab, A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berezovski, M.; Drabovich, A.; Krylova, S.M.; Musheev, M.; Okhonin, V.; Petrov, A.A.; Krylov, S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures: A universal tool for development of aptamers. J. Am. Chem. Soc. 2005, 127, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Meyer, S.; Propson, N.E.; Nie, J.; Jiang, P.; Stewart, R.; Thomson, J. Characterization and target identification of a DNA aptamer that labels pluripotent stem cells. Cell Res. 2015, 25, 390–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for improving aptamer binding affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Shin, K.; Patterson, R.E.; Liu, X.-Q.; Rainey, J.K. Current strategies for protein production and purification enabling membrane protein structural biology. Biochem. Cell Biol. 2016, 94, 507–527. [Google Scholar] [CrossRef] [Green Version]
- Sligar, S.G.; Denisov, I.G. Nanodiscs: A toolkit for membrane protein science. Protein Sci. 2021, 30, 297–315. [Google Scholar] [CrossRef] [PubMed]
- Linnane, E.; Davey, P.; Zhang, P.; Puri, S.; Edbrooke, M.; Chiarparin, E.; Revenko, A.S.; MacLeod, A.R.; Norman, J.C.; Ross, S.J. Differential uptake, kinetics and mechanisms of intracellular trafficking of next-generation antisense oligonucleotides across human cancer cell lines. Nucleic Acids Res. 2019, 47, 4375–4392. [Google Scholar] [CrossRef] [PubMed]




| ID | Random Region Sequence (N30, 5′ to 3′) | Copies in Round 9 | Kd (nM) | T (Utrp)Count | |
|---|---|---|---|---|---|
| TROP2 FcHis | FcHis | ||||
| Tac-A1 | TCTTACCGTTTCCTCGTGCCTTGTTTTCGC | 0.82% | 103 ± 11 | 804 ± 98.8 | 14 |
| Tac-A2 | TCGTTCCTGTTGTGTTCCCTTCTCCTCTGT | 0.62% | 74 ± 8.4 | >1000 | 15 |
| Tac-A3 | TCGTTTACTGTTCCCCTCCTCCTTCCCTTT | 0.43% | 100 ± 7.9 | >1000 | 14 |
| Tac-A4 | TTGTTCCCCCTTTGCCTTTCTTTCCCCTCT | 0.40% | 72 ± 5.4 | >1000 | 15 |
| Tac-A5 | TCTGTTCCGTGTTCGTTCCTTTCCTTGTTG | 0.37% | 50 ± 6.9 | >1000 | 16 |
| ID | Random Region Sequence (N30, 5′ to 3′) | Copies in Round 9 (in Rd 9b) | Kd (nM) | T (Utrp)Count | |
|---|---|---|---|---|---|
| TROP2 FcHis | FcHis | ||||
| Tac-B1 | TGCTGTTGTCACCTGCCTCGTCTCCCTCGT | 0.16% (0.35%) | 153 ± 13 | >1000 | 11 |
| Tac-B2 | TTCCCTCCTCTGTTGTTCCCCCCTCCTCTC | 0.03% (0.32%) | 89 ± 6.0 | >1000 | 12 |
| Tac-B3 | TGGGGTGGTGGTGTGGGTGGGGGTTTGTTC | 0.25% (0.17%) | 187 ± 8.4 | >1000 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uemachi, H.; Kasahara, Y.; Tanaka, K.; Okuda, T.; Yoneda, Y.; Obika, S. Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity. Pharmaceutics 2021, 13, 888. https://doi.org/10.3390/pharmaceutics13060888
Uemachi H, Kasahara Y, Tanaka K, Okuda T, Yoneda Y, Obika S. Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity. Pharmaceutics. 2021; 13(6):888. https://doi.org/10.3390/pharmaceutics13060888
Chicago/Turabian StyleUemachi, Hiro, Yuuya Kasahara, Keisuke Tanaka, Takumi Okuda, Yoshihiro Yoneda, and Satoshi Obika. 2021. "Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity" Pharmaceutics 13, no. 6: 888. https://doi.org/10.3390/pharmaceutics13060888
APA StyleUemachi, H., Kasahara, Y., Tanaka, K., Okuda, T., Yoneda, Y., & Obika, S. (2021). Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity. Pharmaceutics, 13(6), 888. https://doi.org/10.3390/pharmaceutics13060888