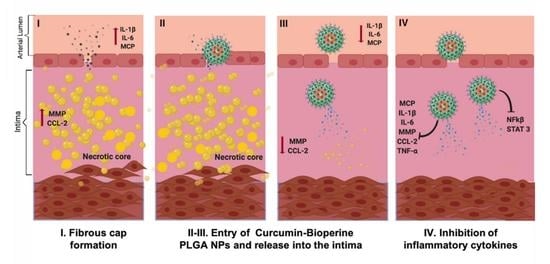
Co-Delivery of Curcumin and Bioperine via PLGA Nanoparticles to Prevent Atherosclerotic Foam Cell Formation
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Synthesis of Curcumin-Bioperine Loaded PLGA Nanoparticles (Cur-Bio-PLGA NPs)
2.3. Yield and Encapsulation Studies
2.4. Characterization of Nanoparticles
2.5. In Vitro Drug Release Studies
2.6. Cell Culture
2.7. THP-1 Cell Differentiation and Foam Cell Formation
2.8. In Vitro Cell Viability Studies
2.9. Cholesterol Quantitation Assay
2.10. Inhibition on Foam Cell Formation by Confocal Microscopy
2.11. Cellular Uptake of Dil-AcLDL by Flow Cytometry
2.12. Quantitative Real-Time PCR (qRT-PCR)
2.13. Statistical Analysis
3. Results and Discussion
3.1. Characterization of Cur-Bio PLGA NPs
3.2. In Vitro Curcumin and Bioperine Release from the NPs
3.3. In Vitro Cell Viability Studies
3.4. Cholesterol Quantitation Assay
3.5. Inhibition on Foam Cell Formation by Confocal Microscopy
3.6. Cellular Uptake of Dil-AcLDL by Flow Cytometry
3.7. Gene Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tabas, I.; García-Cardeña, G.; Owens, G.K. Recent insights into the cellular biology of atherosclerosis. J. Cell Biol. 2015, 209, 13–22. [Google Scholar] [CrossRef]
- Hansson, G.K. Inflammation, Atherosclerosis, and Coronary Artery Disease. N. Engl. J. Med. 2005, 352, 1685–1695. [Google Scholar] [CrossRef] [Green Version]
- Libby, P.; Everett, B.M. Atherosclerosis. Nat. Rev. Dis. Prim. 2019, 5, 56. [Google Scholar] [CrossRef]
- Randolph, G.J. Mechanisms That Regulate Macrophage Burden in Atherosclerosis. Circ. Res. 2014, 114, 1757–1771. [Google Scholar] [CrossRef] [Green Version]
- Linton, M.F.; Yancey, P.G.; Davies, S.S.; Jerome, W.G.; Linton, E.F.; Song, W.L.; Doranm, A.C.; Vickers, K.C. The Role of Lipids and Lipoproteins in Atherosclerosis. 2000. Available online: https://www.ncbi.nlm.nih.gov/books/NBK343489/ (accessed on 3 January 2019).
- Lusis, A.J. Atherosclerosis. Nature 2000, 407, 233–241. [Google Scholar] [CrossRef]
- Li, A.C.; Glass, C.K. The macrophage foam cell as a target for therapeutic intervention. Nat. Med. 2002, 8, 1235–1242. [Google Scholar] [CrossRef]
- Kunjathoor, V.V.; Febbraio, M.; Podrez, E.A.; Moore, K.; Andersson, L.; Koehn, S.; Rhee, J.S.; Silverstein, R.; Hoff, H.F.; Freeman, M.W. Scavenger Receptors Class A-I/II and CD36 Are the Principal Receptors Responsible for the Uptake of Modified Low Density Lipoprotein Leading to Lipid Loading in Macrophages. J. Biol. Chem. 2002, 277, 49982–49988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Febbraio, M.; Podrez, E.A.; Smith, J.D.; Hajjar, D.P.; Hazen, S.L.; Hoff, H.F.; Sharma, K.; Silverstein, R.L. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J. Clin. Investig. 2000, 105, 1049–1056. [Google Scholar] [CrossRef] [Green Version]
- Libby, P. Inflammation in atherosclerosis. Nature 2002, 420, 868–874. [Google Scholar] [CrossRef]
- Dong, Z.M.; Chapman, S.M.; Brown, A.A.; Frenette, P.S.; Hynes, R.O.; Wagner, D.D. The combined role of P- and E-selectins in atherosclerosis. J. Clin. Investig. 1998, 102, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Collins, T.; Cybulsky, M.I. NF-κB: Pivotal mediator or innocent bystander in atherogenesis? J. Clin. Investig. 2001, 107, 255–264. [Google Scholar] [CrossRef]
- Libby, P. Inflammation in Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2045–2051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Sureda, A.; Devkota, H.P.; Pittalà, V.; Barreca, D.; Silva, A.S.; Tewari, D.; Xu, S.; Nabavi, S.M. Curcumin, the golden spice in treating cardiovascular diseases. Biotechnol. Adv. 2020, 38, 107343. [Google Scholar] [CrossRef]
- Luisa, D.P.-A.M.; Griselda, R.-M.; Valentín, M.-L.; Carmina, O.-S.; Cristina, V.-M.; Jj, M.; Maykel, G.-T.; David, Q.-G.; Roberto, S.-S.; Gerardo, L.-G. Curcumin-loaded poly-ε-caprolactone nanoparticles show antioxidant and cytoprotective effects in the presence of reactive oxygen species. J. Bioact. Compat. Polym. 2020, 35, 270–285. [Google Scholar] [CrossRef]
- Joe, B.; Vijaykumar, M.; Lokesh, B.R. Biological Properties of Curcumin-Cellular and Molecular Mechanisms of Action. Crit. Rev. Food Sci. Nutr. 2004, 44, 97–111. [Google Scholar] [CrossRef]
- Chen, Q.; Lv, J.; Yang, W.; Xu, B.; Wang, Z.; Yu, Z.; Wu, J.; Yang, Y.; Han, Y. Targeted inhibition of STAT3 as a potential treatment strategy for atherosclerosis. Theranostics 2019, 9, 6424–6442. [Google Scholar] [CrossRef]
- Liu, T.; Li, C.; Sun, H.; Luo, T.; Tan, Y.; Tian, D.; Guo, Z. Curcumin inhibits monocyte chemoattractant protein-1 expression and enhances cholesterol efflux by suppressing the c-Jun N-terminal kinase pathway in macrophage. Inflamm. Res. 2014, 63, 841–850. [Google Scholar] [CrossRef]
- Bisht, S.; Feldmann, G.; Soni, S.; Ravi, R.; Karikar, C.; Maitra, A.; Maitra, A. Polymeric nanoparticle-encapsulated curcumin ("nanocurcumin"): A novel strategy for human cancer therapy. J. Nanobiotechnol. 2007, 5, 3–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yallapu, M.M.; Jaggi, M.; Chauhan, S.C. Curcumin nanomedicine: A road to cancer therapeutics. Curr. Pharm. Des. 2013, 19, 1994–2010. [Google Scholar] [PubMed]
- Yavarpour-Bali, H.; Ghasemi-Kasman, M.; Pirzadeh, M. Curcumin-loaded nanoparticles: A novel therapeutic strategy in treatment of central nervous system disorders. Int. J. Nanomed. 2019, ume 14, 4449–4460. [Google Scholar] [CrossRef] [Green Version]
- Del Prado-Audelo, M.L.; Caballero-Florán, I.H.; Meza-Toledo, J.A.; Mendoza-Muñoz, N.; González-Torres, M.; Florán, B.; Cortés, H.; Leyva-Gómez, G. Formulations of Curcumin Nanoparticles for Brain Diseases. Biomolecules 2019, 9, 56. [Google Scholar] [CrossRef] [Green Version]
- Borah, A.; Pillai, S.C.; Rochani, A.K.; Palaninathan, V.; Nakajima, Y.; Maekawa, T.; Kumar, D.S. GANT61 and curcumin-loaded PLGA nanoparticles for GLI1 and PI3K/Akt-mediated inhibition in breast adenocarcinoma. Nanotechnology 2020, 31, 185102. [Google Scholar] [CrossRef]
- Sivakumar, B.; Aswathy, R.G.; Nagaoka, Y.; Iwai, S.; Venugopal, K.; Kato, K.; Yoshida, Y.; Maekawa, T.; Kumar, S. Aptamer conjugated theragnostic multifunctional magnetic nanoparticles as a nanoplatform for pancreatic cancer therapy. RSC Adv. 2013, 3, 20579–20598. [Google Scholar] [CrossRef]
- Mathew, A.; Fukuda, T.; Nagaoka, Y.; Hasumura, T.; Morimoto, H.; Yoshida, Y.; Maekawa, T.; Venugopal, K.; Kumar, D.S. Curcumin Loaded-PLGA Nanoparticles Conjugated with Tet-1 Peptide for Potential Use in Alzheimer’s Disease. PLoS ONE 2012, 7, e32616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, N.; Gong, Y.; Zhang, J.; Mu, X.; Song, Z.; Feng, R.; Zhang, H. A novel curcumin-loaded nanoparticle restricts atherosclerosis development and promotes plaques stability in apolipoprotein E deficient mice. J. Biomater. Appl. 2018, 33, 946–954. [Google Scholar] [CrossRef]
- Randhawa, G.K.; Kullar, J.S. Rajkumar Bioenhancers from mother nature and their applicability in modern medicine. Int. J. Appl. Basic Med. Res. 2011, 1, 5–10. [Google Scholar] [CrossRef]
- Shoba, G.; Joy, D.; Joseph, T.; Majeed, M.; Rajendran, R.; Srinivas, P.S.S.R. Influence of Piperine on the Pharmacokinetics of Curcumin in Animals and Human Volunteers. Planta Medica 1998, 64, 353–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atal, N.; Bedi, K. Bioenhancers: Revolutionary concept to market. J. Ayurveda Integr. Med. 2010, 1, 96–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baspinar, Y.; Üstündas, M.; Bayraktar, O.; Sezgin, C. Curcumin and piperine loaded zein-chitosan nanoparticles: Development and in-vitro characterisation. Saudi Pharm. J. 2018, 26, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Budama-Kilinc, Y. Piperine Nanoparticles for Topical Application: Preparation, Characterization, In vitro and In silico Evaluation. ChemistrySelect 2019, 4, 11693–11700. [Google Scholar] [CrossRef]
- Bhalekar, M.R.; Madgulkar, A.R.; Desale, P.S.; Marium, G. Formulation of piperine solid lipid nanoparticles (SLN) for treatment of rheumatoid arthritis. Drug Dev. Ind. Pharm. 2017, 43, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.C.; Borah, A.; Jindal, A.; Jacob, E.M.; Yamamoto, Y.; Kumar, D.S. BioPerine Encapsulated Nanoformulation for Overcoming Drug-Resistant Breast Cancers. Asian J. Pharm. Sci. 2020, 15, 701–712. [Google Scholar] [CrossRef]
- Pachauri, M.; Gupta, E.D.; Ghosh, P.C. Piperine loaded PEG-PLGA nanoparticles: Preparation, characterization and targeted delivery for adjuvant breast cancer chemotherapy. J. Drug Deliv. Sci. Technol. 2015, 29, 269–282. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of Curcumin: Problems and Promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Makadia, H.K.; Siegel, S.J. Poly Lactic-co-Glycolic Acid (PLGA) as Biodegradable Controlled Drug Delivery Carrier. Polymers 2011, 3, 1377–1397. [Google Scholar] [CrossRef]
- Sun, S.-B.; Liu, P.; Shao, F.-M.; Miao, Q.-L. Formulation and evaluation of PLGA nanoparticles loaded capecitabine for prostate cancer. Int. J. Clin. Exp. Med. 2015, 8, 19670–19681. [Google Scholar]
- Borah, A.; Palaninathan, V.; Girija, A.R.; Balasubramanian, S.; Rochani, A.K.; Maekawa, T.; Kumar, D.S. Poly-lactic-co-glycolic acid Nanoformulation of Small Molecule Antagonist GANT61 for Cancer Annihilation by Modulating Hedgehog Pathway. NanoWorld J. 2017, 3. [Google Scholar] [CrossRef]
- Takashiba, S.; Van Dyke, T.E.; Amar, S.; Murayama, Y.; Soskolne, A.W.; Shapira, L. Differentiation of monocytes to macrophages primes cells for lipopolysaccharide stimulation via accumulation of cytoplasmic nuclear factor kappaB. Infect. Immun. 1999, 67, 5573–5578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, T.K.; Bostanian, L.A.; Graves, R.A.; Chapman, S.R. Poly(D,L-lactide-co-glycolide) encapsulated poly(vinyl alcohol) hydrogel as a drug delivery system. Pharm. Res. 2002, 19, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Erbetta, C.D.C.; Alves, R.J.; Resende, J.M.; Freitas, R.F.d.S.; Sous, R.G.d. Synthesis and Characterization of Poly(D,L-Lactide-co-Glycolide) Copolymer. J. Biomater. Nanobiotechnol. 2012, 3, 208–225. [Google Scholar] [CrossRef]
- Darandale, S.; Vavia, P.R. Cyclodextrin-based nanosponges of curcumin: Formulation and physicochemical characterization. J. Incl. Phenom. Macrocycl. Chem. 2012, 75, 315–322. [Google Scholar] [CrossRef]
- Gorgani, L.; Mohammadi, M.; Nikzad, M.; Najafpour, G.D. Sequential Microwave-Ultrasound-Assisted Extraction for Isolation of Piperine from Black Pepper (Piper nigrum L.). Food Bioprocess Technol. 2017, 10, 2199–2207. [Google Scholar] [CrossRef]
- Shaikh, J.; Ankola, D.; Beniwal, V.; Singh, D.; Kumar, M.R. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur. J. Pharm. Sci. 2009, 37, 223–230. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, B.; Wang, Y.; Lou, D. Dual Drug Release from Core–Shell Nanoparticles with Distinct Release Profiles. J. Pharm. Sci. 2014, 103, 3205–3216. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Nie, L.; Zou, P.; Suo, J. Effects of drug and polymer molecular weight on drug release from PLGA-mPEG microspheres. J. Appl. Polym. Sci. 2015, 132, 41431. [Google Scholar] [CrossRef]
- Hasan, M.; ElKhoury, K.; Kahn, C.; Arab-Tehrany, E.; Linder, M. Preparation, Characterization, and Release Kinetics of Chitosan-Coated Nanoliposomes Encapsulating Curcumin in Simulated Environments. Molecules 2019, 24, 2023. [Google Scholar] [CrossRef] [Green Version]
- Prabhuraj, R.S.; Bomb, K.; Srivastava, R.; Bandyopadhyaya, R. Dual drug delivery of curcumin and niclosamide using PLGA nanoparticles for improved therapeutic effect on breast cancer cells. J. Polym. Res. 2020, 27, 1–13. [Google Scholar] [CrossRef]
- Kamaly, N.; Yameen, B.; Wu, J.; Farokhzad, O.C. Degradable Controlled-Release Polymers and Polymeric Nanoparticles: Mechanisms of Controlling Drug Release. Chem. Rev. 2016, 116, 2602–2663. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.-K.; Ha, T.-Y.; McGregor, R.A.; Choi, M.-S. Long-term curcumin administration protects against atherosclerosis via hepatic regulation of lipoprotein cholesterol metabolism. Mol. Nutr. Food Res. 2011, 55, 1829–1840. [Google Scholar] [CrossRef]
- Min, K.; Um, H.J.; Cho, K.-H.; Kwon, T.K. Curcumin inhibits oxLDL-induced CD36 expression and foam cell formation through the inhibition of p38 MAPK phosphorylation. Food Chem. Toxicol. 2013, 58, 77–85. [Google Scholar] [CrossRef]
- Peschel, D.; Koerting, R.; Nass, N. Curcumin induces changes in expression of genes involved in cholesterol homeostasis. J. Nutr. Biochem. 2007, 18, 113–119. [Google Scholar] [CrossRef]
- Zingg, J.-M.; Hasan, S.T.; Cowan, D.; Ricciarelli, R.; Azzi, A.; Meydani, M. Regulatory effects of curcumin on lipid accumulation in monocytes/macrophages. J. Cell. Biochem. 2012, 113, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-F.; Ching, L.-C.; Huang, Y.-C.; Chen, C.-Y.; Chiang, A.-N.; Kou, Y.R.; Shyue, S.-K.; Lee, T.-S. Molecular mechanism of curcumin on the suppression of cholesterol accumulation in macrophage foam cells and atherosclerosis. Mol. Nutr. Food Res. 2012, 56, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Tannock, L.R. Advances in the management of hyperlipidemia-induced atherosclerosis. Expert Rev. Cardiovasc. Ther. 2008, 6, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Feng, J.; Fan, Z.; Li, J. Curcumin increases cholesterol efflux via heme oxygenase-1-mediated ABCA1 and SR-BI expression in macrophages. Mol. Med. Rep. 2018, 17, 6138–6143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.-L.; Liu, M.-H.; Hu, H.-J.; Feng, H.-R.; Fan, X.-J.; Zou, W.-W.; Pan, Y.-Q.; Hu, X.-M.; Wang, Z. Curcumin Enhanced Cholesterol Efflux by Upregulating ABCA1 Expression Through AMPK-SIRT1-LXRα Signaling in THP-1 Macrophage-Derived Foam Cells. DNA Cell Biol. 2015, 34, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.B.; Mengi, S.A.; Xu, Y.-J.; Arneja, A.S.; Dhalla, N.S. Pathogenesis of atherosclerosis: A multifactorial process. Exp. Clin. Cardiol. 2002, 7, 40–53. [Google Scholar]
- Pober, J.S.; Sessa, W. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 2007, 7, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.D.; Elias, M.G.; Dellinger, A.L.; Kepley, C.L. NF-κB inhibitors that prevent foam cell formation and atherosclerotic plaque accumulation. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2037–2048. [Google Scholar] [CrossRef]
- Hasan, S.; Zingg, J.-M.; Kwan, P.; Noble, T.; Smith, D.; Meydani, M. Curcumin modulation of high fat diet-induced atherosclerosis and steatohepatosis in LDL receptor deficient mice. Atherosclerosis 2014, 232, 40–51. [Google Scholar] [CrossRef]
- Phillips, M.C. Molecular Mechanisms of Cellular Cholesterol Efflux. J. Biol. Chem. 2014, 289, 24020–24029. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Curcumin-Bioperine PLGA Nanoparticles | Particle Size (nm) | PDI a | ZP b (mV) | % EE c | % YE d | % LE e | ||
|---|---|---|---|---|---|---|---|---|
| Curcumin | Bioperine | Curcumin | Bioperine | |||||
| 1:1:10 | 293 ± 4.15 | 0.01 ± 0.004 | −21 ± 0.88 | 97.4 ± 0.44 | 98.2 ± 0.03 | 55 ± 1.73 | 16.25 ± 0.3 | 15.67 ± 0.68 |
| 1:0.2:10 | 181 ± 8.63 | 0.21 ± 0.005 | −17.6 ± 0.33 | 98.2 ± 1.01 | 85.8 ± 0.41 | 49.6 ± 0.88 | 17.9 ± 1.2 | 17.2 ± 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pillai, S.C.; Borah, A.; Le, M.N.T.; Kawano, H.; Hasegawa, K.; Kumar, D.S. Co-Delivery of Curcumin and Bioperine via PLGA Nanoparticles to Prevent Atherosclerotic Foam Cell Formation. Pharmaceutics 2021, 13, 1420. https://doi.org/10.3390/pharmaceutics13091420
Pillai SC, Borah A, Le MNT, Kawano H, Hasegawa K, Kumar DS. Co-Delivery of Curcumin and Bioperine via PLGA Nanoparticles to Prevent Atherosclerotic Foam Cell Formation. Pharmaceutics. 2021; 13(9):1420. https://doi.org/10.3390/pharmaceutics13091420
Chicago/Turabian StylePillai, Sindhu C., Ankita Borah, Minh Nguyen Tuyet Le, Hiroaki Kawano, Kouichi Hasegawa, and D. Sakthi Kumar. 2021. "Co-Delivery of Curcumin and Bioperine via PLGA Nanoparticles to Prevent Atherosclerotic Foam Cell Formation" Pharmaceutics 13, no. 9: 1420. https://doi.org/10.3390/pharmaceutics13091420
APA StylePillai, S. C., Borah, A., Le, M. N. T., Kawano, H., Hasegawa, K., & Kumar, D. S. (2021). Co-Delivery of Curcumin and Bioperine via PLGA Nanoparticles to Prevent Atherosclerotic Foam Cell Formation. Pharmaceutics, 13(9), 1420. https://doi.org/10.3390/pharmaceutics13091420