Comparative Analysis of Free-Circulating and Vesicle-Associated Plasma microRNAs of Healthy Controls and Early-Stage Lung Cancer Patients
Abstract
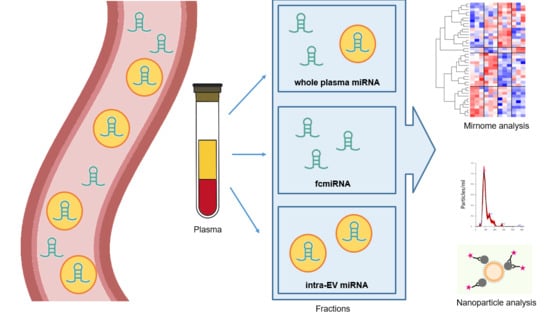
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. EV Isolation and RNA Extraction
2.3. Nanoparticle Analysis
2.4. Protein Analysis of EV lysates
2.5. microRNA Profiling
2.6. Bioinformatic and Statistical Analysis
3. Results
3.1. Difference of RNA and Library Profiles Obtained by Different Extraction Methods
3.2. Sequencing Performance According to Extraction Method
3.3. Different miRNA Expression Profiles Identified According to Extraction Method
3.4. FC and EV-Associated microRNA Profiles in Lung Cancer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wong, S.Q.; Dawson, S.-J. Combining Liquid Biopsies and PET-CT for Early Cancer Detection. Nat. Med. 2020, 26, 1010–1011. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B. Liquid Biopsy versus Tumor Biopsy for Clinical-Trial Recruitment. Nat. Med. 2020, 26, 1815–1816. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical Relevance of Blood-Based CtDNA Analysis: Mutation Detection and Beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Herbst, R.S.; Boshoff, C. Toward Personalized Treatment Approaches for Non-Small-Cell Lung Cancer. Nat. Med. 2021, 27, 1345–1356. [Google Scholar] [CrossRef]
- Im, Y.R.; Tsui, D.W.Y.; Diaz, L.A.; Wan, J.C.M. Next-Generation Liquid Biopsies: Embracing Data Science in Oncology. Trends Cancer 2020, 7, 283–292. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Inoue, J.; Inazawa, J. Cancer-Associated MiRNAs and Their Therapeutic Potential. J. Hum. Genet. 2021, 66, 937–945. [Google Scholar] [CrossRef]
- Moon, S.; Shin, D.W.; Kim, S.; Lee, Y.-S.; Mankhong, S.; Yang, S.W.; Lee, P.H.; Park, D.-H.; Kwak, H.-B.; Lee, J.-S.; et al. Enrichment of Exosome-Like Extracellular Vesicles from Plasma Suitable for Clinical Vesicular MiRNA Biomarker Research. J. Clin. Med. 2019, 8, 1995. [Google Scholar] [CrossRef]
- Zickler, A.M.; Andaloussi, S.E. Functional Extracellular Vesicles Aplenty. Nat. Biomed. Eng. 2020, 4, 9–11. [Google Scholar] [CrossRef]
- Endzeliņš, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Soboļevska, K.; Ābols, A.; Rodriguez, M.; Šantare, D.; Rudņickiha, A.; Lietuvietis, V.; et al. Detection of Circulating MiRNAs: Comparative Analysis of Extracellular Vesicle-Incorporated MiRNAs and Cell-Free MiRNAs in Whole Plasma of Prostate Cancer Patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef]
- Hulstaert, E.; Morlion, A.; Cobos, F.A.; Verniers, K.; Nuytens, J.; Eynde, E.V.; Yigit, N.; Anckaert, J.; Geerts, A.; Hindryckx, P.; et al. Charting Extracellular Transcriptomes in The Human Biofluid RNA Atlas. Cell Rep. 2020, 33, 108552. [Google Scholar] [CrossRef] [PubMed]
- Peña-Llopis, S.; Brugarolas, J. Simultaneous Isolation of High-Quality DNA, RNA, MiRNA and Proteins from Tissues for Genomic Applications. Nat. Protoc. 2013, 8, 2240–2255. [Google Scholar] [CrossRef] [PubMed]
- Notarangelo, M.; Zucal, C.; Modelska, A.; Pesce, I.; Scarduelli, G.; Potrich, C.; Lunelli, L.; Pederzolli, C.; Pavan, P.; Marca, G.L.; et al. Ultrasensitive Detection of Cancer Biomarkers by Nickel-Based Isolation of Polydisperse Extracellular Vesicles from Blood. eBiomedicine 2019, 43, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid Biopsy Enters the Clinic—Implementation Issues and Future Challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.K.Y.; MacMahon, M.; Woodside, J.V.; Simpson, D.A. A Comparison of RNA Extraction and Sequencing Protocols for Detection of Small RNAs in Plasma. BMC Genom. 2019, 20, 446. [Google Scholar] [CrossRef] [PubMed]
- Kloten, V.; Neumann, M.H.D.; Pasquale, F.D.; Sprenger-Haussels, M.; Shaffer, J.M.; Schlumpberger, M.; Herdean, A.; Betsou, F.; Ammerlaan, W.; Hällström, T.A.; et al. Multicenter Evaluation of Circulating Plasma MicroRNA Extraction Technologies for the Development of Clinically Feasible Reverse Transcription Quantitative PCR and Next-Generation Sequencing Analytical Work Flows. Clin. Chem. 2019, 65, 1132–1140. [Google Scholar] [CrossRef]
- Yeri, A.; Courtright, A.; Danielson, K.; Hutchins, E.; Alsop, E.; Carlson, E.; Hsieh, M.; Ziegler, O.; Das, A.; Shah, R.V.; et al. Evaluation of Commercially Available Small RNASeq Library Preparation Kits Using Low Input RNA. BMC Genom. 2018, 19, 331. [Google Scholar] [CrossRef]
- Coenen-Stass, A.M.L.; Magen, I.; Brooks, T.; Ben-Dov, I.Z.; Greensmith, L.; Hornstein, E.; Fratta, P. Evaluation of Methodologies for MicroRNA Biomarker Detection by next Generation Sequencing. RNA Biol. 2018, 15, 1133–1145. [Google Scholar] [CrossRef]
- Schneegans, S.; Lück, L.; Besler, K.; Bluhm, L.; Stadler, J.; Staub, J.; Greinert, R.; Volkmer, B.; Kubista, M.; Gebhardt, C.; et al. Pre-analytical Factors Affecting the Establishment of a Single Tube Assay for Multiparameter Liquid Biopsy Detection in Melanoma Patients. Mol. Oncol. 2020, 14, 1001–1015. [Google Scholar] [CrossRef]
- Alsop, E.; Meechoovet, B.; Kitchen, R.; Sweeney, T.; Beach, T.G.; Serrano, G.E.; Hutchins, E.; Ghiran, I.; Reiman, R.; Syring, M.; et al. A Novel Tissue Atlas and Online Tool for the Interrogation of Small RNA Expression in Human Tissues and Biofluids. Front. Cell Dev. Biol. 2022, 10, 804164. [Google Scholar] [CrossRef]
- Takov, K.; Yellon, D.M.; Davidson, S.M. Comparison of Small Extracellular Vesicles Isolated from Plasma by Ultracentrifugation or Size-Exclusion Chromatography: Yield, Purity and Functional Potential. J. Extracell. Vesicles 2018, 8, 1560809. [Google Scholar] [CrossRef] [PubMed]
- Reale, A.; Carmichael, I.; Xu, R.; Mithraprabhu, S.; Khong, T.; Chen, M.; Fang, H.; Savvidou, I.; Ramachandran, M.; Bingham, N.; et al. Human Myeloma Cell- and Plasma-derived Extracellular Vesicles Contribute to Functional Regulation of Stromal Cells. Proteomics 2021, 21, 2000119. [Google Scholar] [CrossRef] [PubMed]
- Arioz, B.I.; Tufekci, K.U.; Olcum, M.; Durur, D.Y.; Akarlar, B.A.; Ozlu, N.; Bagriyanik, H.A.; Keskinoglu, P.; Yener, G.; Genc, S. Proteome Profiling of Neuron-Derived Exosomes in Alzheimer’s Disease Reveals Hemoglobin as a Potential Biomarker. Neurosci. Lett. 2021, 755, 135914. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Cucchiari, D.; Tubita, V.; Rovira, J.; Ramirez-Bajo, M.J.; Banon-Maneus, E.; Lazo-Rodriguez, M.; Hierro-Garcia, N.; Borràs, F.E.; Ventura-Aguiar, P.; Piñeiro, G.J.; et al. B Cell-Derived Extracellular Vesicles Reveal Residual B Cell Activity in Kidney Graft Recipients Undergoing Pre-Transplant Desensitization. Front. Med. 2021, 8, 781239. [Google Scholar] [CrossRef]
- Wu, J.; Wu, H.; An, J.; Ballantyne, C.M.; Cyster, J.G. Critical Role of Integrin CD11c in Splenic Dendritic Cell Capture of Missing-Self CD47 Cells to Induce Adaptive Immunity. Proc. Nat. Acad. Sci. USA 2018, 115, 6786–6791. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef]
- Lee, S.-S.; Won, J.-H.; Lim, G.J.; Han, J.; Lee, J.Y.; Cho, K.-O.; Bae, Y.-K. A Novel Population of Extracellular Vesicles Smaller than Exosomes Promotes Cell Proliferation. Cell Commun. Signal. 2019, 17, 95. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Cheng, L.; Hill, A.F. Small RNA Library Construction for Exosomal RNA from Biological Samples for the Ion Torrent PGM™ and Ion S5™ System. Methods Mol. Biol. 2017, 1545, 71–90. [Google Scholar] [CrossRef]
- Androvic, P.; Benesova, S.; Rohlova, E.; Kubista, M.; Valihrach, L. Small RNA-Sequencing for Analysis of Circulating MiRNAs. J. Mol. Diagn. 2022, 24, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Heinicke, F.; Zhong, X.; Zucknick, M.; Breidenbach, J.; Sundaram, A.Y.M.; Flåm, S.T.; Leithaug, M.; Dalland, M.; Farmer, A.; Henderson, J.M.; et al. Systematic Assessment of Commercially Available Low-Input MiRNA Library Preparation Kits. RNA Biol. 2019, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Dama, E.; Melocchi, V.; Mazzarelli, F.; Colangelo, T.; Cuttano, R.; Di Candia, L.; Ferretti, G.M.; Taurchini, M.; Graziano, P.; Bianchi, F. Non-Coding RNAs as Prognostic Biomarkers: A miRNA Signature Specific for Aggressive Early-Stage Lung Adenocarcinomas. Noncoding RNA 2020, 6, 48. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, H.; Gao, X.; Wei, F.; Zhang, X.; Su, Y.; Wang, C.; Li, H.; Ren, X. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget 2016, 7, 26070-86. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qin, F.; Hu, F.; Xu, H.; Sun, G.; Han, G.; Wang, T.; Guo, M. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018, 8, 2. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, H.; Zhang, L.; Zhou, X.; Wang, T.; Zhang, J.; Shu, Y.; Zhu, W.; Wen, W.; Liu, P. Identification of four plasma microRNAs as potential biomarkers in the diagnosis of male lung squamous cell carcinoma patients in China. Cancer Med. 2018, 7, 2370–2381. [Google Scholar] [CrossRef]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef]
- Sun, Y.; Mei, H.; Xu, C.; Tang, H.; Wei, W. Circulating microRNA-339-5p and -21 in plasma as an early detection predictors of lung adenocarcinoma. Pathol. Res. Pract. 2018, 214, 119–125. [Google Scholar] [CrossRef]
- Zhong, S.; Golpon, H.; Zardo, P.; Borlak, J. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. Res. 2021, 230, 164–196. [Google Scholar] [CrossRef]
- Italiano, J.E.; Mairuhu, A.T.; Flaumenhaft, R. Clinical Relevance of Microparticles from Platelets and Megakaryocytes. Curr. Opin. Hematol. 2010, 17, 578–584. [Google Scholar] [CrossRef] [Green Version]
- Flaumenhaft, R.; Mairuhu, A.T.A.; Italiano, J.E. Platelet- and Megakaryocyte-Derived Microparticles. Semin. Thromb. Hemost. 2010, 36, 881–887. [Google Scholar] [CrossRef]
- Yang, W.; Yao, Y.; Chen, Y.; Jin, F.; Zheng, T.; Ai, X.; Zhang, T.; Ding, C.; Jiang, M. Dynamic Changes of Platelets before and after Surgery Predict the Prognosis of Patients with Operable Non-Small Cell Lung Cancer. J. Cancer 2022, 13, 823–830. [Google Scholar] [CrossRef]
- Chen, C.; Yang, H.; Cai, D.; Xiang, L.; Fang, W.; Wang, R. Preoperative Peripheral Blood Neutrophil-to-Lymphocyte Ratios (NLR) and Platelet-to-Lymphocyte Ratio (PLR) Related Nomograms Predict the Survival of Patients with Limited-Stage Small-Cell Lung Cancer. Transl. Lung. Cancer Res. 2021, 10, 866. [Google Scholar] [CrossRef]
- Wang, H.; Li, C.; Yang, R.; Jin, J.; Liu, D.; Li, W. Prognostic Value of the Platelet-to-Lymphocyte Ratio in Lung Cancer Patients Receiving Immunotherapy: A Systematic Review and Meta-Analysis. PLoS ONE 2022, 17, e0268288. [Google Scholar] [CrossRef] [PubMed]





| Sample | Gender | Age | Histological Diagnosis 1 | Stage | Smoking History |
|---|---|---|---|---|---|
| L01 | M | 72 | ADC | IB | yes |
| L02 | F | 75 | ADC | IIA | yes |
| L03 | F | 71 | ADC | IIIA | yes |
| L04 | F | 68 | ADC | IA | yes |
| L05 | M | 70 | ADC | IIB | yes |
| L06 | M | 71 | SQC | IIIA | yes |
| L07 | F | 72 | ADC | IIIA | yes |
| L08 | M | 77 | SQC | IB | yes |
| L09 | M | 55 | ADC | IA | yes |
| L10 | F | 66 | ADC | IB | no |
| L11 | M | 79 | SQC | IIIA | yes |
| L12 | F | 70 | SQC | IIB | yes |
| L13 | F | 55 | ADC | IA | yes |
| L14 | M | 83 | ADC | IB | no |
| L15 | M | 55 | SQC | IIIA | yes |
| L16 | M | 72 | ADC | IB | yes |
| L17 | M | 69 | ADC | IIIA | yes |
| L18 | M | 72 | ADC | IIIA | yes |
| RNA Fraction | Name | Hits | Pop Hits | p Value | Adj. p Value |
|---|---|---|---|---|---|
| WP | GO:0005844_polysome | 12 | 42 | 0.0 | 0.0 |
| WP | GO:0030122_AP-2_adaptor_complex | 10 | 19 | 0.0 | 0.0 |
| WP | GO:0032587_ruffle_membrane | 18 | 105 | 0.0001 | 0.0056 |
| WP | GO:0000792_heterochromatin | 11 | 44 | 0.0002 | 0.0084 |
| WP | GO:0005845_mRNA_cap_binding_complex | 6 | 14 | 0.0005 | 0.0092 |
| WP | GO:0005925_focal_adhesion | 50 | 498 | 0.0004 | 0.0092 |
| WP | GO:0008021_synaptic_vesicle | 20 | 134 | 0.0003 | 0.0092 |
| WP | GO:0016442_RISC_complex | 7 | 20 | 0.0005 | 0.0092 |
| WP | GO:0016581_NuRD_complex | 7 | 21 | 0.0006 | 0.0092 |
| WP | GO:0016607_nuclear_speck | 45 | 443 | 0.0006 | 0.0092 |
| secEV | GO:0005758_mitochondrial_intermembrane | 12 | 98 | 0.0003 | 0.0127 |
| secEV | GO:0005759_mitochondrial_matrix | 31 | 447 | 0.0004 | 0.0127 |
| secEV | GO:0014069_postsynaptic_density | 22 | 270 | 0.0003 | 0.0127 |
| secEV | GO:0005942_PI3K_complex | 5 | 25 | 0.0026 | 0.0494 |
| secEV | GO:0043235_receptor_complex | 19 | 257 | 0.0022 | 0.0494 |
| ccEV | GO:0005802_trans-Golgi_network | 9 | 207 | 0.0008 | 0.0132 |
| ccEV | GO:0055037_recycling_endosome | 8 | 146 | 0.0004 | 0.0132 |
| ccEV | GO:0005770_late_endosome | 7 | 157 | 0.0026 | 0.0214 |
| ccEV | GO:0005925_focal_adhesion | 14 | 498 | 0.0024 | 0.0214 |
| ccEV | GO:0005776_autophagosome | 5 | 91 | 0.0044 | 0.029 |
| ccEV | GO:0000139_Golgi_membrane | 16 | 682 | 0.0066 | 0.0311 |
| ccEV | GO:0030667_secretory_granule_membrane | 5 | 100 | 0.0064 | 0.0311 |
| ccEV | GO:0005769_early_endosome | 9 | 301 | 0.0087 | 0.0359 |
| FC | GO:0005769_early_endosome | 31 | 301 | 0.0 | 0.0 |
| FC | GO:0031901_early_endosome_membrane | 20 | 173 | 0.0 | 0.0 |
| FC | GO:0071141_SMAD_protein_complex | 5 | 8 | 0.0 | 0.0 |
| FC | GO:1990124_messenger_ribonucleoprotein_comp | 5 | 11 | 0.0001 | 0.0025 |
| FC | GO:0000139_Golgi_membrane | 44 | 682 | 0.0002 | 0.004 |
| FC | GO:0016363_nuclear_matrix | 12 | 112 | 0.001 | 0.0156 |
| FC | GO:0055037_recycling_endosome | 14 | 146 | 0.0011 | 0.0156 |
| FC | GO:0030014_CCR4-NOT_complex | 5 | 20 | 0.0013 | 0.0161 |
| FC | GO:0035098_ESCE(Z)_complex | 5 | 23 | 0.0022 | 0.0242 |
| FC | GO:0005635_nuclear_envelope | 17 | 217 | 0.0027 | 0.0264 |
| RNA Fraction | Name | Hits | Pop Hits | p Value | Adj. p Value |
|---|---|---|---|---|---|
| FC | GO:0000123_histone_acetyltransferase_complex | 7 | 27 | 0.0003 | 0.0119 |
| FC | GO:0030122_AP-2_adaptor_complex | 6 | 19 | 0.0003 | 0.0119 |
| FC | GO:0031209_SCAR_complex | 6 | 17 | 0.0002 | 0.0119 |
| FC | GO:0017146_NMDA_selective_glutamate_recept. | 6 | 21 | 0.0005 | 0.0149 |
| FC | GO:0032591_dendritic_spine_membrane | 5 | 15 | 0.0008 | 0.0159 |
| FC | GO:0090575_RNA_polymerase_II_transcription | 11 | 80 | 0.0007 | 0.0159 |
| FC | GO:0010494_cytoplasmic_stress_granule | 11 | 84 | 0.0011 | 0.0187 |
| FC | GO:0071782_endoplasmic_reticulum_tubular_nt | 5 | 20 | 0.0023 | 0.0342 |
| FC | GO:0098982_GABA-ergic_synapse | 9 | 69 | 0.003 | 0.0397 |
| FC | GO:0045211_postsynaptic_membrane | 14 | 146 | 0.0037 | 0.044 |
| ccEV | GO:0000159_protein_phosphatase_type2A_compl. | 7 | 32 | 0.0 | 0.0 |
| ccEV | GO:0032593_insulin-responsive_compartment | 5 | 13 | 0.0 | 0.0 |
| ccEV | GO:0010494_cytoplasmic_stress_granule | 9 | 84 | 0.03 | 1.11 |
| ccEV | GO:0032991_protein-containing_complex | 32 | 679 | 0.04 | 1.11 |
| ccEV | GO:0000792_heterochromatin | 6 | 44 | 0.09 | 2.08 |
| ccEV | GO:0036464_cytoplasmic_ribonucleoprot_granule | 8 | 85 | 0.13 | 2.34 |
| ccEV | GO:0035097_histone_methyltransferase_complex | 5 | 33 | 0.16 | 2.42 |
| ccEV | GO:0000307_CDK_holoenzyme_complex | 5 | 35 | 0.21 | 3.06 |
| ccEV | GO:0000932_P-body | 9 | 116 | 0.24 | 3.09 |
| ccEV | GO:0030027_lamellipodium | 12 | 198 | 0.36 | 4.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasini, L.; Vannini, I.; Ulivi, P.; Tebaldi, M.; Petracci, E.; Fabbri, F.; Stella, F.; Urbini, M. Comparative Analysis of Free-Circulating and Vesicle-Associated Plasma microRNAs of Healthy Controls and Early-Stage Lung Cancer Patients. Pharmaceutics 2022, 14, 2029. https://doi.org/10.3390/pharmaceutics14102029
Pasini L, Vannini I, Ulivi P, Tebaldi M, Petracci E, Fabbri F, Stella F, Urbini M. Comparative Analysis of Free-Circulating and Vesicle-Associated Plasma microRNAs of Healthy Controls and Early-Stage Lung Cancer Patients. Pharmaceutics. 2022; 14(10):2029. https://doi.org/10.3390/pharmaceutics14102029
Chicago/Turabian StylePasini, Luigi, Ivan Vannini, Paola Ulivi, Michela Tebaldi, Elisabetta Petracci, Francesco Fabbri, Franco Stella, and Milena Urbini. 2022. "Comparative Analysis of Free-Circulating and Vesicle-Associated Plasma microRNAs of Healthy Controls and Early-Stage Lung Cancer Patients" Pharmaceutics 14, no. 10: 2029. https://doi.org/10.3390/pharmaceutics14102029
APA StylePasini, L., Vannini, I., Ulivi, P., Tebaldi, M., Petracci, E., Fabbri, F., Stella, F., & Urbini, M. (2022). Comparative Analysis of Free-Circulating and Vesicle-Associated Plasma microRNAs of Healthy Controls and Early-Stage Lung Cancer Patients. Pharmaceutics, 14(10), 2029. https://doi.org/10.3390/pharmaceutics14102029