Biomaterial-Mediated Protein Expression Induced by Peptide-mRNA Nanoparticles Embedded in Lyophilized Collagen Scaffolds
Abstract
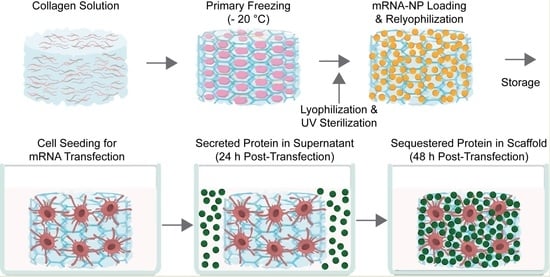
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Media
2.2. Preparation of Collagen Scaffolds
2.3. Messenger RNA (mRNA)
2.4. Formation of Transfection Complexes
2.5. Nanoparticle (NP) Diffusion into Collagen Scaffolds
2.6. Optimization of the Cell Seeding Density
2.7. Presence of Viable Cells 24 h Post-Seeding
2.8. Determination of Pre-Lyophilization Collagen Scaffold Volume
2.9. Dose–Response Profile of PF14-SecNLuc mRNA-Loaded Collagen Scaffolds
2.10. Effects of Storage Temperature on Luciferase mRNA Transfections
2.11. In Vitro BMP-7 Production in BMP-7 mRNA-Loaded Collagen Scaffolds
2.12. Data Analysis and Statistics
3. Results
3.1. mRNA Nanoparticles Distribute Homogeneously throughout a 3D Collagen Scaffold after Lyophilization
3.2. Cell Seeding Density and Pre-Lyophilization Volume of Collagen Scaffolds Are Crucial Parameters for In Vitro Cell Viability
3.3. C2C12 and MC3T3 Cells Decorate Collagen Fibers throughout a 3D Scaffold
3.4. Peptide-Mediated mRNA Delivery in 3D Collagen Scaffolds Induces Dose-Dependent Protein Production
3.5. Long-Term Storage of mRNA-Loaded Collagen Scaffolds at Different Temperatures Decreases Transfection Efficiencies
3.6. BMP-7 mRNA Transfection in Collagen Scaffolds Results in Biomaterial-Mediated Protein Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 161. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2020, 384, 403–416. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [Green Version]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Klauer, A.A.; van Hoof, A. Degradation of mRNAs that lack a stop codon: A decade of nonstop progress. Wiley Interdiscip. Rev. RNA 2012, 3, 649–660. [Google Scholar] [CrossRef] [Green Version]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [Green Version]
- Yokoo, H.; Oba, M.; Uchida, S. Cell-Penetrating Peptides: Emerging Tools for mRNA Delivery. Pharmaceutics 2021, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Reesor, E.K.G.; Xu, Y.; Zope, H.R.; Zetter, B.R.; Shi, J. Biomaterials for mRNA delivery. Biomater. Sci. 2015, 3, 1519–1533. [Google Scholar] [CrossRef] [Green Version]
- Fenton, O.S.; Olafson, K.N.; Pillai, P.S.; Mitchell, M.J.; Langer, R. Advances in Biomaterials for Drug Delivery. Adv. Mater. 2018, e1705328. [Google Scholar] [CrossRef] [PubMed]
- Andrée, L.; Yang, F.; Brock, R.; Leeuwenburgh, S.C.G. Designing biomaterials for the delivery of RNA therapeutics to stimulate bone healing. Mater. Today Bio 2021, 10, 100105. [Google Scholar] [CrossRef]
- Damiati, L.A.; El-Messeiry, S. An Overview of RNA-Based Scaffolds for Osteogenesis. Front. Mol. Biosci. 2021, 8, 454. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, G.S.; Roffi, A.; Reale, D.; Kon, E.; Filardo, G. Clinical application of bone morphogenetic proteins for bone healing: A systematic review. Int. Orthop. 2017, 41, 1073–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreira, A.C.; Lojudice, F.H.; Halcsik, E.; Navarro, R.D.; Sogayar, M.C.; Granjeiro, J.M. Bone Morphogenetic Proteins: Facts, Challenges, and Future Perspectives. J. Dent. Res. 2014, 93, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.T.; Rosenbaum, A.J. Bone grafts, bone substitutes and orthobiologics. Organogenesis 2012, 8, 114–124. [Google Scholar] [CrossRef] [Green Version]
- Epstein, N.E. Pros, cons, and costs of INFUSE in spinal surgery. Surg. Neurol. Int. 2011, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Epstein, N.E. Complications due to the use of BMP/INFUSE in spine surgery: The evidence continues to mount. Surg. Neurol. Int. 2013, 4, S343–S352. [Google Scholar] [CrossRef]
- Poon, B.; Kha, T.; Tran, S.; Dass, C.R. Bone morphogenetic protein-2 and bone therapy: Successes and pitfalls. J. Pharm. Pharmacol. 2016, 68, 139–147. [Google Scholar] [CrossRef]
- Vandermeer, J.S.; Kamiya, N.; Aya-ay, J.; Garces, A.; Browne, R.; Kim, H.K.W. Local administration of ibandronate and bone morphogenetic protein-2 after ischemic osteonecrosis of the immature femoral head: A combined therapy that stimulates bone formation and decreases femoral head deformity. J. Bone Jt. Surg. Am. 2011, 93, 905–913. [Google Scholar] [CrossRef]
- Kim, H.K.W.; Aruwajoye, O.; Du, J.; Kamiya, N. Local administration of bone morphogenetic protein-2 and bisphosphonate during non-weight-bearing treatment of ischemic osteonecrosis of the femoral head: An experimental investigation in immature pigs. J. Bone Jt. Surg. Am. 2014, 96, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.S.; La, W.-G.; Cho, Y.-M.; Shin, W.; Yeo, G.-D.; Kim, B.-S. Comparison between heparin-conjugated fibrin and collagen sponge as bone morphogenetic protein-2 carriers for bone regeneration. Exp. Mol. Med. 2012, 44, 350–355. [Google Scholar] [CrossRef] [Green Version]
- De La Vega, R.E.; van Griensven, M.; Zhang, W.; Coenen, M.J.; Nagelli, C.V.; Panos, J.A.; Peniche Silva, C.J.; Geiger, J.; Plank, C.; Evans, C.H.; et al. Efficient healing of large osseous segmental defects using optimized chemically modified messenger RNA encoding BMP-2. Sci. Adv. 2022, 8, eabl6242. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.A.; Alba-Perez, A.; Gonzalez-Garcia, C.; Donnelly, H.; Llopis-Hernandez, V.; Jayawarna, V.; Childs, P.; Shields, D.W.; Cantini, M.; Ruiz-Cantu, L.; et al. Nanoscale Coatings for Ultralow Dose BMP-2-Driven Regeneration of Critical-Sized Bone Defects. Adv. Sci. 2019, 6, 1800361. [Google Scholar] [CrossRef] [PubMed]
- Rivera, J.; Strohbach, C.; Wenke, J.; Rathbone, C. Beyond osteogenesis: An in vitro comparison of the potentials of six bone morphogenetic proteins. Front. Pharmacol. 2013, 4, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillman, C.E.; Jayasuriya, A.C. FDA-approved bone grafts and bone graft substitute devices in bone regeneration. Mater. Sci. Eng. C 2021, 130, 112466. [Google Scholar] [CrossRef]
- Vukicevic, S.; Oppermann, H.; Verbanac, D.; Jankolija, M.; Popek, I.; Curak, J.; Brkljacic, J.; Pauk, M.; Erjavec, I.; Francetic, I.; et al. The clinical use of bone morphogenetic proteins revisited: A novel biocompatible carrier device OSTEOGROW for bone healing. Int. Orthop. 2014, 38, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Giannoudis, P.V.; Tzioupis, C. Clinical applications of BMP-7: The UK perspective. Injury 2005, 36 (Suppl. 3), S47–S50. [Google Scholar] [CrossRef]
- Granjeiro, J.M.; Oliveira, R.C.; Bustos-Valenzuela, J.C.; Sogayar, M.C.; Taga, R. Bone morphogenetic proteins: From structure to clinical use. Braz. J. Med. Biol. Res. 2005, 38, 1463–1473. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.C.; Maitra, S.; Anderson, M.J.; Christiansen, B.A.; Reddi, A.H.; Lee, M.A. BMP-7 and Bone Regeneration: Evaluation of Dose-Response in a Rodent Segmental Defect Model. J. Orthop. Trauma 2015, 29, e336-41. [Google Scholar] [CrossRef]
- Cecchi, S.; Bennet, S.J.; Arora, M. Bone morphogenetic protein-7: Review of signalling and efficacy in fracture healing. J. Orthop. Transl. 2015, 4, 28–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaccaro, A.R.; Whang, P.G.; Patel, T.; Phillips, F.M.; Anderson, D.G.; Albert, T.J.; Hilibrand, A.S.; Brower, R.S.; Kurd, M.F.; Appannagari, A.; et al. The safety and efficacy of OP-1 (rhBMP-7) as a replacement for iliac crest autograft for posterolateral lumbar arthrodesis: Minimum 4-year follow-up of a pilot study. Spine J. 2008, 8, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, J.W.; Blizzard, D.J. The controversy surrounding bone morphogenetic proteins in the spine: A review of current research. Yale J. Biol. Med. 2014, 87, 549–561. [Google Scholar] [PubMed]
- Badieyan, Z.S.; Berezhanskyy, T.; Utzinger, M.; Aneja, M.K.; Emrich, D.; Erben, R.; Schüler, C.; Altpeter, P.; Ferizi, M.; Hasenpusch, G.; et al. Transcript-activated collagen matrix as sustained mRNA delivery system for bone regeneration. J. Control. Release 2016, 239, 137–148. [Google Scholar] [CrossRef]
- Elangovan, S.; Khorsand, B.; Do, A.-V.; Hong, L.; Dewerth, A.; Kormann, M.; Ross, R.D.; Rick Sumner, D.; Allamargot, C.; Salem, A.K. Chemically modified RNA activated matrices enhance bone regeneration. J. Control. Release 2015, 218, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; De La Vega, R.E.; Coenen, M.J.; Müller, S.A.; Peniche Silva, C.J.; Aneja, M.K.; Plank, C.; van Griensven, M.; Evans, C.H.; Balmayor, E.R. An Improved, Chemically Modified RNA Encoding BMP-2 Enhances Osteogenesis In Vitro and In Vivo. Tissue Eng. Part A 2018, 25, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Duan, H.; Xu, L.; Witman, N.; Yan, B.; Yu, Z.; Wang, H.; Tan, Y.; Lin, L.; Li, D.; et al. BMP-2 and VEGF-A modRNAs in collagen scaffold synergistically drive bone repair through osteogenic and angiogenic pathways. Commun. Biol. 2021, 4, 82. [Google Scholar] [CrossRef]
- Balmayor, E.R.; Geiger, J.P.; Koch, C.; Aneja, M.K.; van Griensven, M.; Rudolph, C.; Plank, C. Modified mRNA for BMP-2 in Combination with Biomaterials Serves as a Transcript-Activated Matrix for Effectively Inducing Osteogenic Pathways in Stem Cells. Stem Cells Dev. 2017, 26, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Perche, F.; Midoux, P.; Cabral, C.S.; Cátia, S.D.; Malard, V.; Correia, I.J.; EI-Hafci, H.; Petite, H.; Logeart-Avramoglou, D.; et al. In Vivo bone tissue induction by freeze-dried collagen-nanohydroxyapatite matrix loaded with BMP2/NS1 mRNAs lipopolyplexes. J. Control. Release 2021, 334, 188–200. [Google Scholar] [CrossRef]
- Khalil, A.S.; Yu, X.; Umhoefer, J.M.; Chamberlain, C.S.; Wildenauer, L.A.; Diarra, G.M.; Hacker, T.A.; Murphy, W.L. Single-dose mRNA therapy via biomaterial-mediated sequestration of overexpressed proteins. Sci. Adv. 2020, 6, eaba2422. [Google Scholar] [CrossRef]
- van den Brand, D.; Gorris, M.A.J.; van Asbeck, A.H.; Palmen, E.; Ebisch, I.; Dolstra, H.; Hällbrink, M.; Massuger, L.F.A.G.; Brock, R. Peptide-mediated delivery of therapeutic mRNA in ovarian cancer. Eur. J. Pharm. Biopharm. 2019, 141, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Faraj, K.A.; van Kuppevelt, T.H.; Daamen, W.F. Construction of collagen scaffolds that mimic the three-dimensional architecture of specific tissues. Tissue Eng. 2007, 13, 2387–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brouwer, K.M.; Wijnen, R.M.; Reijnen, D.; Hafmans, T.G.; Daamen, W.F.; van Kuppevelt, T.H. Heparinized collagen scaffolds with and without growth factors for the repair of diaphragmatic hernia: Construction and in vivo evaluation. Organogenesis 2013, 9, 161–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacio-Castañeda, V.; Oude Egberink, R.; Sait, A.; Andrée, L.; Sala, B.M.; Hassani Besheli, N.; Oosterwijk, E.; Nilvebrant, J.; Leeuwenburgh, S.C.G.; Brock, R.; et al. Mimicking the Biology of Engineered Protein and mRNA Nanoparticle Delivery Using a Versatile Microfluidic Platform. Pharmaceuticals 2021, 13, 1944. [Google Scholar] [CrossRef]
- Ke, M.-T.; Fujimoto, S.; Imai, T. SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013, 16, 1154–1161. [Google Scholar] [CrossRef]
- Preibisch, S.; Saalfeld, S.; Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25, 1463–1465. [Google Scholar] [CrossRef] [PubMed]
- van Asbeck, A.H.; Dieker, J.; Oude Egberink, R.; van den Berg, L.; van der Vlag, J.; Brock, R. Protein Expression Correlates Linearly with mRNA Dose over Up to Five Orders of Magnitude In Vitro and In Vivo. Biomedicines 2021, 9, 511. [Google Scholar] [CrossRef]
- Sork, H.; Nordin, J.Z.; Turunen, J.J.; Wiklander, O.P.B.; Bestas, B.; Zaghloul, E.M.; Margus, H.; Padari, K.; Duru, A.D.; Corso, G.; et al. Lipid-based Transfection Reagents Exhibit Cryo-induced Increase in Transfection Efficiency. Mol. Ther. Nucleic Acids 2016, 5, e290. [Google Scholar] [CrossRef] [Green Version]
- Husteden, C.; Doberenz, F.; Goergen, N.; Pinnapireddy, S.R.; Janich, C.; Langner, A.; Syrowatka, F.; Repanas, A.; Erdmann, F.; Jedelská, J.; et al. Contact-Triggered Lipofection from Multilayer Films Designed as Surfaces for in Situ Transfection Strategies in Tissue Engineering. ACS Appl. Mater. Interfaces 2020, 12, 8963–8977. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, H.; Yan, J.; Bryers, J.D. Scaffold-mediated delivery for non-viral mRNA vaccines. Gene Ther. 2018, 25, 556–567. [Google Scholar] [CrossRef]
- Ball, R.L.; Bajaj, P.; Whitehead, K.A. Achieving long-term stability of lipid nanoparticles: Examining the effect of pH, temperature, and lyophilization. Int. J. Nanomed. 2016, 12, 305–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Hou, X.; Yan, J.; Du, S.; Xue, Y.; Li, W.; Xiang, G.; Dong, Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact. Mater. 2020, 5, 358–363. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Mesquida, P.; Kohl, D.; Andriotis, O.G.; Thurner, P.J.; Duer, M.; Bansode, S.; Schitter, G. Evaluation of surface charge shift of collagen fibrils exposed to glutaraldehyde. Sci. Rep. 2018, 8, 10126. [Google Scholar] [CrossRef]
- Migliorini, E.; Guevara-Garcia, A.; Albiges-Rizo, C.; Picart, C. Learning from BMPs and their biophysical extracellular matrix microenvironment for biomaterial design. Bone 2020, 141, 115540. [Google Scholar] [CrossRef] [PubMed]
- van Asbeck, A.H.; Beyerle, A.; McNeill, H.; Bovee-Geurts, P.H.M.; Lindberg, S.; Verdurmen, W.P.R.; Hällbrink, M.; Langel, Ü.; Heidenreich, O.; Brock, R. Molecular Parameters of siRNA–Cell Penetrating Peptide Nanocomplexes for Efficient Cellular Delivery. ACS Nano 2013, 7, 3797–3807. [Google Scholar] [CrossRef]
- Yen, A.; Cheng, Y.; Sylvestre, M.; Gustafson, H.H.; Puri, S.; Pun, S.H. Serum Nuclease Susceptibility of mRNA Cargo in Condensed Polyplexes. Mol. Pharm. 2018, 15, 2268–2276. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef]
- Sturm, L.; Schwemberger, B.; Menzel, U.; Häckel, S.; Albers, C.E.; Plank, C.; Rip, J.; Alini, M.; Traweger, A.; Grad, S.; et al. In Vitro Evaluation of a Nanoparticle-Based mRNA Delivery System for Cells in the Joint. Biomedicines 2021, 9, 794. [Google Scholar] [CrossRef]
- Ramsay, E.; Gumbleton, M. Polylysine and Polyornithine Gene Transfer Complexes: A Study of Complex Stability and Cellular Uptake as a Basis for their Differential in-vitro Transfection Efficiency. J. Drug Target. 2002, 10, 1–9. [Google Scholar] [CrossRef]
- Ezzat, K.; Andaloussi, S.E.L.; Zaghloul, E.M.; Lehto, T.; Lindberg, S.; Moreno, P.M.D.; Viola, J.R.; Magdy, T.; Abdo, R.; Guterstam, P.; et al. PepFect 14, a novel cell-penetrating peptide for oligonucleotide delivery in solution and as solid formulation. Nucleic Acids Res. 2011, 39, 5284–5298. [Google Scholar] [CrossRef] [PubMed]
- Dirisala, A.; Uchida, S.; Li, J.; Van Guyse, J.F.R.; Hayashi, K.; Vummaleti, S.V.C.; Kaur, S.; Mochida, Y.; Fukushima, S.; Kataoka, K. Effective mRNA Protection by Poly(l-ornithine) Synergizes with Endosomal Escape Functionality of a Charge-Conversion Polymer toward Maximizing mRNA Introduction Efficiency. Macromol. Rapid Commun. 2022, 43, e2100754. [Google Scholar] [CrossRef]
- Semple, S.C.; Harasym, T.O.; Clow, K.A.; Ansell, S.M.; Klimuk, S.K.; Hope, M.J. Immunogenicity and Rapid Blood Clearance of Liposomes Containing Polyethylene Glycol-Lipid Conjugates and Nucleic Acid. J. Pharmacol. Exp. Ther. 2005, 312, 1020–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogueira, S.S.; Schlegel, A.; Maxeiner, K.; Weber, B.; Barz, M.; Schroer, M.A.; Blanchet, C.E.; Svergun, D.I.; Ramishetti, S.; Peer, D.; et al. Polysarcosine-Functionalized Lipid Nanoparticles for Therapeutic mRNA Delivery. ACS Appl. Nano Mater. 2020, 3, 10634–10645. [Google Scholar] [CrossRef]
- Schlapschy, M.; Binder, U.; Börger, C.; Theobald, I.; Wachinger, K.; Kisling, S.; Haller, D.; Skerra, A. PASylation: A biological alternative to PEGylation for extending the plasma half-life of pharmaceutically active proteins. Protein Eng. Des. Sel. 2013, 26, 489–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Packer, M.; Gyawali, D.; Yerabolu, R.; Schariter, J.; White, P. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat. Commun. 2021, 12, 6777. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oude Egberink, R.; Zegelaar, H.M.; El Boujnouni, N.; Versteeg, E.M.M.; Daamen, W.F.; Brock, R. Biomaterial-Mediated Protein Expression Induced by Peptide-mRNA Nanoparticles Embedded in Lyophilized Collagen Scaffolds. Pharmaceutics 2022, 14, 1619. https://doi.org/10.3390/pharmaceutics14081619
Oude Egberink R, Zegelaar HM, El Boujnouni N, Versteeg EMM, Daamen WF, Brock R. Biomaterial-Mediated Protein Expression Induced by Peptide-mRNA Nanoparticles Embedded in Lyophilized Collagen Scaffolds. Pharmaceutics. 2022; 14(8):1619. https://doi.org/10.3390/pharmaceutics14081619
Chicago/Turabian StyleOude Egberink, Rik, Helen M. Zegelaar, Najoua El Boujnouni, Elly M. M. Versteeg, Willeke F. Daamen, and Roland Brock. 2022. "Biomaterial-Mediated Protein Expression Induced by Peptide-mRNA Nanoparticles Embedded in Lyophilized Collagen Scaffolds" Pharmaceutics 14, no. 8: 1619. https://doi.org/10.3390/pharmaceutics14081619
APA StyleOude Egberink, R., Zegelaar, H. M., El Boujnouni, N., Versteeg, E. M. M., Daamen, W. F., & Brock, R. (2022). Biomaterial-Mediated Protein Expression Induced by Peptide-mRNA Nanoparticles Embedded in Lyophilized Collagen Scaffolds. Pharmaceutics, 14(8), 1619. https://doi.org/10.3390/pharmaceutics14081619