Conjugation of Short Oligopeptides to a Second-Generation Polyamidoamine Dendrimer Shows Antibacterial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis and Characterization of FKFL-G2 PAMAM Dendrimer
2.3. Characterization of the FKFL-G2
2.3.1. Mass Spectrometry
2.3.2. Critical Aggregation Concentration (CAC)
2.3.3. Zeta Potential and Size
2.4. Cytotoxicity Assay
2.5. Bacterial Growth Assay
2.6. Colony-Forming Unit Assay
2.7. Minimum Growth Inhibitory Concentration (MIC) Test
2.8. Membrane Permeabilization Assay
2.9. Scanning Electron Microscopy
2.10. Biofilm Formation Assay
2.11. Statistical Analysis
3. Results
3.1. Characterization of the FKFL-G2 Dendrimer
3.2. Cytotoxicity Assay
3.3. Bacterial Growth Assay
3.4. Colony-Forming Unit Assay
3.5. Membrane Permeabilization Assay
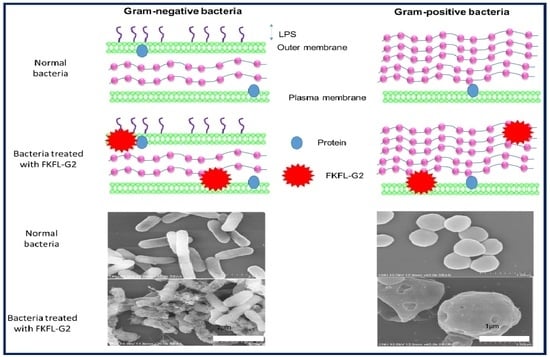
3.6. Scanning Electron Microscopy
3.7. Biofilm Formation Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmad, M.; Khan, A.U. Global economic impact of antibiotic resistance: A review. J. Glob. Antimicrob. Resist. 2019, 19, 313–316. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [Green Version]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [Green Version]
- Araújo, R.; Santos, S.; Ferreira, E.I.; Giarolla, J. New Advances in General Biomedical Applications of PAMAM Dendrimers. Molecules 2018, 23, 2849. [Google Scholar] [CrossRef] [Green Version]
- Cong, H.; Zhou, L.; Meng, Q.; Zhang, Y.; Yu, B.; Shen, Y.; Hu, H. Preparation and evaluation of PAMAM dendrimer-based polymer gels physically cross-linked by hydrogen bonding. Biomater. Sci. 2019, 7, 3918–3925. [Google Scholar] [CrossRef]
- Labieniec-Watala, M.; Watala, C. PAMAM Dendrimers: Destined for Success or Doomed to Fail? Plain and Modified PAMAM Dendrimers in the Context of Biomedical Applications. J. Pharm. Sci. 2015, 104, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Majoros, I.J.; Myc, A.; Thomas, T.; Mehta, C.B.; Baker, J.R. PAMAM Dendrimer-Based Multifunctional Conjugate for Cancer Therapy: Synthesis, Characterization, and Functionality. Biomacromolecules 2006, 7, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Li, D.; Shi, X.; Shen, M. PAMAM Dendrimer-Based Nanodevices for Nuclear Medicine Applications. Macromol. Biosci. 2019, 20, e1900282. [Google Scholar] [CrossRef] [PubMed]
- Esumi, K.; Houdatsu, H.; Yoshimura, T. Antioxidant Action by Gold−PAMAM Dendrimer Nanocomposites. Langmuir 2004, 20, 2536–2538. [Google Scholar] [CrossRef]
- Huh, A.J.; Kwon, Y.J. “Nanoantibiotics”: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 2011, 156, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Arteta, M.Y.; Ainalem, M.-L.; Porcar, L.; Martel, A.; Coker, H.; Lundberg, D.; Chang, D.P.; Soltwedel, O.; Barker, R.; Nylander, T. Interactions of PAMAM Dendrimers with Negatively Charged Model Biomembranes. J. Phys. Chem. B 2014, 118, 12892–12906. [Google Scholar] [CrossRef] [PubMed]
- Fox, L.J.; Richardson, R.M.; Briscoe, W.H. PAMAM dendrimer-cell membrane interactions. Adv. Colloid Interface Sci. 2018, 257, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Huang, R.; Liu, S.; Huang, S.; Jiang, C. Peptide-Conjugated PAMAM for Targeted Doxorubicin Delivery to Transferrin Receptor Overexpressed Tumors. Mol. Pharm. 2010, 7, 2156–2165. [Google Scholar] [CrossRef]
- Xu, X.; Li, J.; Han, S.; Tao, C.; Fang, L.; Sun, Y.; Zhu, J.; Liang, Z.; Li, F. A novel doxorubicin loaded folic acid conjugated PAMAM modified with borneol, a nature dual-functional product of reducing PAMAM toxicity and boosting BBB penetration. Eur. J. Pharm. Sci. 2016, 88, 178–190. [Google Scholar] [CrossRef]
- Waite, C.L.; Roth, C.M. PAMAM-RGD Conjugates Enhance siRNA Delivery Through a Multicellular Spheroid Model of Malignant Glioma. Bioconjugate Chem. 2009, 20, 1908–1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kharwade, R.; More, S.; Warokar, A.; Agrawal, P.; Mahajan, N. Starburst pamam dendrimers: Synthetic approaches, surface modifications, and biomedical applications. Arab. J. Chem. 2020, 13, 6009–6039. [Google Scholar] [CrossRef]
- Calabretta, M.K.; Kumar, A.; McDermott, A.M.; Cai, C. Antibacterial Activities of Poly(amidoamine) Dendrimers Terminated with Amino and Poly(ethylene glycol) Groups. Biomacromolecules 2007, 8, 1807–1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, A.; Reins, R.; McDermott, A.; Trautner, B.; Cai, C. Antibacterial activity and cytotoxicity of PEGylated poly (am-idoamine) dendrimers. MolBiosyst 2009, 5, 1148–1156. [Google Scholar]
- Winnicka, K.; Wroblewska, M.; Wieczorek, P.; Sacha, P.; Tryniszewska, E. The effect of PAMAM dendrimers on the anti-bacterial activity of antibiotics with different water solubility. Molecules 2013, 18, 8607–8617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serri, A.; Mahboubi, A.; Zarghi, A.; Moghimi, H. PAMAM-dendrimer enhanced antibacterial effect of vancomycin hydrochlo-ride against gram-negative bacteria. Int. J. Pharm. Pharm. Sci. 2019, 22, 10–21. [Google Scholar]
- Cheng, Y.; Qu, H.; Ma, M.; Xu, Z.; Xu, P.; Fang, Y.; Xu, T. Polyamidoamine (PAMAM) dendrimers as biocompatible carriers of quinolone antimicrobials: An in vitro study. Eur. J. Med. Chem. 2007, 42, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Gholami, M.; Mohammadi, R.; Arzanlou, M.; Dourbash, F.A.; Kouhsari, E.; Majidi, G.; Mohseni, S.; Nazari, S. In vitro an-tibacterial activity of poly (amidoamine)-G7 dendrimer. BMC Infect. Dis. 2017, 17, 395. [Google Scholar] [CrossRef] [Green Version]
- Holmes, A.; Heylings, J.; Wan, K.-W.; Moss, G. Antimicrobial efficacy and mechanism of action of poly (amidoam-ine)(PAMAM) dendrimers against opportunistic pathogens. Int. J. Antimicrob. Agents 2019, 53, 500–507. [Google Scholar] [CrossRef] [PubMed]
- SMatboo, A.; Nazari, S.; Niapour, A.; Niri, M.; Asgari, E.; Mokhtari, S. Antibacterial effect of TiO2 modified with poly-amidoamine dendrimer–G3 on S. aureus and E. coli in aqueous solutions. Water Sci. Technol. 2022, 85, 605–616. [Google Scholar] [CrossRef]
- Gou, Y.; Yang, X.; He, L.; Xu, X.; Liu, Y.; Liu, Y.; Gao, Y.; Huang, Q.; Liang, K.; Ding, C.; et al. Bio-inspired peptide decorated dendrimers for a robust antibacterial coating on hydroxyapatite. Polym. Chem. 2017, 8, 4264–4279. [Google Scholar] [CrossRef]
- ABahar, A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar]
- Valenti, G.E.; Alfei, S.; Caviglia, D.; Domenicotti, C.; Marengo, B. Antimicrobial Peptides and Cationic Nanoparticles: A Broad-Spectrum Weapon to Fight Multi-Drug Resistance Not Only in Bacteria. Int. J. Mol. Sci. 2022, 23, 6108. [Google Scholar] [CrossRef]
- Hirano, M.; Saito, C.; Yokoo, H.; Goto, C.; Kawano, R.; Misawa, T.; Demizu, Y. Development of Antimicrobial Stapled Peptides Based on Magainin 2 Sequence. Molecules 2021, 26, 444. [Google Scholar] [CrossRef]
- Tam, J.; Wang, S.; Wong, K.; Tan, W. Antimicrobial peptides from plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Tsuda, N.; Miyata, T.; Ikenaga, M. Antimicrobial effect and mechanism of bovine lactoferrin against the potato common scab pathogen Streptomyces scabiei. PLoS ONE 2022, 17, e0264094. [Google Scholar] [CrossRef]
- Cavallo-Medved, D.; Moin, K.; Sloane, B. Cathepsin B: Basis Sequence: Mouse. AFCS Nat. Mol. Pages 2011, 2011, A000508. [Google Scholar]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta BBA-Proteins Proteom. 2012, 1824, 68–88. [Google Scholar] [CrossRef] [Green Version]
- Mort, J.; Buttle, D. Cathepsin b. Int. J. Biochem. Cell Biol. 1997, 29, 715–720. [Google Scholar] [CrossRef]
- Gondi, C.S.; Rao, J.S. Cathepsin B as a cancer target. Expert Opin. Ther. Targets 2013, 17, 281–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szulc-Dąbrowska, L.; Bossowska-Nowicka, M.; Struzik, J.; Toka, F.N. Cathepsins in Bacteria-Macrophage Interaction: Defenders or Victims of Circumstance? Front. Cell. Infect. Microbiol. 2020, 10, 601072. [Google Scholar] [CrossRef]
- Tong, B.; Wan, B.; Wei, Z.; Wang, T.; Zhao, P.; Dou, Y.; Lv, Z.; Xia, Y.; Dai, Y. Role of cathepsin B in regulating migration and invasion of fibroblast-like synoviocytes into inflamed tissue from patients with rheumatoid arthritis. Clin. Exp. Immunol. 2014, 177, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-E.; Ho, C.-C.; Yang, S.-F.; Lin, S.-H.; Yeh, K.-T.; Lin, C.-W.; Chen, M.-K. Cathepsin B Expression and the Correlation with Clinical Aspects of Oral Squamous Cell Carcinoma. PLoS ONE 2016, 11, e0152165. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Shao, J.; Gao, L.; Qi, Y.; Ye, L. Adsorption of Cathepsin B-sensitive peptide conjugated DOX on nanodiamonds. Appl. Surf. Sci. 2011, 257, 8617–8622. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, J.; Jin, X.; Liu, L.; Tian, X. Folate Receptor Targeting and Cathepsin B-Sensitive Drug Delivery System for Selective Cancer Cell Death and Imaging. ACS Med. Chem. Lett. 2020, 11, 1514–1520. [Google Scholar] [CrossRef]
- Lee, S.; Song, S.J.; Lee, J.; Ha, T.H.; Choi, J.S. Cathepsin B-Responsive Liposomes for Controlled Anticancer Drug Delivery in Hep G2 Cells. Pharmaceutics 2020, 12, 876. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Son, S.; Song, S.; Ha, T.; Choi, J. Polyamidoamine (PAMAM) dendrimers modified with cathepsin-B cleavable oli-gopeptides for enhanced gene delivery. Polymers 2017, 9, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Park, S.; Kwon, Y.-E.; Oh, E.; Kim, D.; Guim, H.; Yeon, J.; Kim, J.-C.; Choi, J. Synthesis and characterization of Du-al-Sensitive PAMAM derivatives conjugated with enzyme cleavable peptides as gene carriers. Macromol. Res. 2021, 29, 636–647. [Google Scholar] [CrossRef]
- Li, L.; Vorobyov, I.; Allen, T.W. The Different Interactions of Lysine and Arginine Side Chains with Lipid Membranes. J. Phys. Chem. B 2013, 117, 11906–11920. [Google Scholar] [CrossRef]
- Winter, C.G.; Christensen, H.N. Migration of Amino Acids across the Membrane of the Human Erythrocyte. J. Biol. Chem. 1964, 239, 872–878. [Google Scholar] [CrossRef]
- Misra, S.; Mukherjee, S.; Ghosh, A.; Singh, P.; Mondal, S.; Ray, D.; Bhattacharya, G.; Ganguly, D.; Ghosh, A.; Aswal, V. Single Amino-Acid Based Self-Assembled Biomaterials with Potent Antimicrobial Activity. Chem. Eur. J. 2021, 27, 16744–16753. [Google Scholar] [CrossRef]
- Kesharwani, A.K.; Mishra, J. Detection of β-lactamase and antibiotic susceptibility of clinical isolates of Staphylococcus aureus. Biocatal. Agric. Biotechnol. 2019, 17, 720–725. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Symonds, P.; Murray, J.C.; Hunter, A.; Debska, G.; Szewczyk, A.C. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2015, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Braissant, O.; Astasov-Frauenhoffer, M.; Waltimo, T.; Bonkat, G. A Review of Methods to Determine Viability, Vitality, and Metabolic Rates in Microbiology. Front. Microbiol. 2020, 11, 547458. [Google Scholar] [CrossRef]
- Yamamura, H.; Hagiwara, T.; Hayashi, Y.; Osawa, K.; Kato, H.; Katsu, T.; Masuda, K.; Sumino, A.; Yamashita, H.; Jinno, R.; et al. Antibacterial Activity of Membrane-Permeabilizing Bactericidal Cyclodextrin Derivatives. ACS Omega 2021, 6, 31831–31842. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by bacterial human pathogens: Clinical relevance-development, composition and regulation-therapeutical strategies. Microb. Cell 2021, 8, 28–56. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar]
- Duncan, R.; Izzo, L. Dendrimer biocompatibility and toxicity. Adv. Drug Deliv. Rev. 2005, 57, 2215–2237. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.; Beuerman, R. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front. Neurosci. 2017, 11, 73. [Google Scholar]
- Lishchynskyi, O.; Shymborska, Y.; Stetsyshyn, Y.; Raczkowska, J.; Skirtach, A.G.; Peretiatko, T.; Budkowski, A. Passive antifouling and active self-disinfecting antiviral surfaces. Chem. Eng. J. 2022, 446, 137048. [Google Scholar] [CrossRef]









| Concentration (µM) | Zeta Potential | Diameter a | Polydispersity (PDI) a |
|---|---|---|---|
| 42.3 | 29.63 ± 6.51 | 158.33 ± 12.59 | 0.628 ± 0.14 |
| 84.6 | 35.5 ± 6.0 | 109.83 ± 17.54 | 0.459 ± 0.18 |
| 169.2 | 38.23 ± 5.74 | 98.45 ± 6.95 | 0.475 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, N.; Thuy, L.T.; Dongquoc, V.; Choi, J.S. Conjugation of Short Oligopeptides to a Second-Generation Polyamidoamine Dendrimer Shows Antibacterial Activity. Pharmaceutics 2023, 15, 1005. https://doi.org/10.3390/pharmaceutics15031005
Kang N, Thuy LT, Dongquoc V, Choi JS. Conjugation of Short Oligopeptides to a Second-Generation Polyamidoamine Dendrimer Shows Antibacterial Activity. Pharmaceutics. 2023; 15(3):1005. https://doi.org/10.3390/pharmaceutics15031005
Chicago/Turabian StyleKang, Namyoung, Le Thi Thuy, Viet Dongquoc, and Joon Sig Choi. 2023. "Conjugation of Short Oligopeptides to a Second-Generation Polyamidoamine Dendrimer Shows Antibacterial Activity" Pharmaceutics 15, no. 3: 1005. https://doi.org/10.3390/pharmaceutics15031005
APA StyleKang, N., Thuy, L. T., Dongquoc, V., & Choi, J. S. (2023). Conjugation of Short Oligopeptides to a Second-Generation Polyamidoamine Dendrimer Shows Antibacterial Activity. Pharmaceutics, 15(3), 1005. https://doi.org/10.3390/pharmaceutics15031005