Influence of Surface Ligand Density and Particle Size on the Penetration of the Blood–Brain Barrier by Porous Silicon Nanoparticles
Abstract
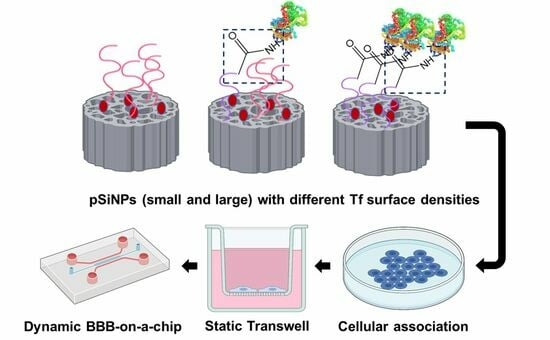
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Attenuated Total Reflection-Fourier Transform Infrared Spectroscopy (ATR-FTIR)
2.2.2. Transmission Electron Microscopy (TEM)
2.2.3. Dynamic Light Scattering (DLS)
2.2.4. Electrochemical Etching of Silicon Wafer
2.2.5. Thermal Hydrocarbonization of Porous Silicon Film
2.2.6. Fractioning of Silicon Wafer by Ultrasonication
2.2.7. Size Selection and Functionalization of pSiNPs with Undecylenic Acid (UDA)
2.2.8. Preparation of Cy5-Labeled Transferrin and PEG-Conjugated Nanoparticles
2.2.9. Cell Culture
2.2.10. Cell Viability in Contact with pSiNPs
2.2.11. Cellular Association and Uptake of pSiNPs via Flow Cytometry and Confocal Microscopy
2.2.12. BBB Transwell Model Preparation for Nanoparticles Assessment
2.2.13. BBB-on-a-Chip Model Establishment and Nanoparticles Assessment
3. Results and Discussions
3.1. Preparation of pSiNPs with Various Ligand Surface Densities and Sizes
3.2. Evaluation of pSiNPs in Brain Microvascular Endothelial Cells
3.3. Assessment of pSiNPs Using Transwell Assay
3.4. Assessment of pSiNPs Using BBB-on-a-Chip Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, C.; Wu, Y.; Chen, X.; Chen, Y.; Wu, Z.; Lin, Z.; Kang, D.; Fang, W.; Chen, F. Global, regional, and national burden and attributable risk factors of neurological disorders: The Global Burden of Disease study 1990–2019. Front. Public Health 2022, 10, 952161. [Google Scholar] [CrossRef]
- Iqbal, K.; Liu, F.; Gong, C.-X. Tau and neurodegenerative disease: The story so far. Nat. Rev. Neurol. 2016, 12, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef]
- Chu, C.; Jablonska, A.; Lesniak, W.G.; Thomas, A.M.; Lan, X.; Linville, R.M.; Li, S.; Searson, P.C.; Liu, G.; Pearl, M.; et al. Optimization of osmotic blood-brain barrier opening to enable intravital microscopy studies on drug delivery in mouse cortex. J. Control. Release 2020, 317, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-K.; Chao, S.-P.; Hu, C.-J. Clinical trials of new drugs for Alzheimer disease. J. Biomed. Sci. 2020, 27, 18. [Google Scholar] [CrossRef]
- Cirotti, C.; Contadini, C.; Barilà, D. SRC Kinase in Glioblastoma: News from an Old Acquaintance. Cancers 2020, 12, 1558. [Google Scholar] [CrossRef]
- Anthony, D.P.; Hegde, M.; Shetty, S.S.; Rafic, T.; Mutalik, S.; Rao, B.S.S. Targeting receptor-ligand chemistry for drug delivery across blood-brain barrier in brain diseases. Life Sci. 2021, 274, 119326. [Google Scholar] [CrossRef]
- Pardridge, W.M. The blood-brain barrier: Bottleneck in brain drug development. NeuroRx 2005, 2, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Swissa, E.; Serlin, Y.; Vazana, U.; Prager, O.; Friedman, A. Blood–brain barrier dysfunction in status epileptics: Mechanisms and role in epileptogenesis. Epilepsy Behav. 2019, 101, 106285. [Google Scholar] [CrossRef]
- Burgess, A.; Shah, K.; Hough, O.; Hynynen, K. Focused ultrasound-mediated drug delivery through the blood–brain barrier. Expert Rev. Neurother. 2015, 15, 477–491. [Google Scholar] [CrossRef]
- Hersh, D.S.; Wadajkar, A.S.; Roberts, N.; Perez, J.G.; Connolly, N.P.; Frenkel, V.; Winkles, J.A.; Woodworth, G.F.; Kim, A.J. Evolving Drug Delivery Strategies to Overcome the Blood Brain Barrier. Curr. Pharm. Des. 2016, 22, 1177–1193. [Google Scholar] [CrossRef]
- Furtado, D.; Björnmalm, M.; Ayton, S.; Bush, A.I.; Kempe, K.; Caruso, F. Overcoming the blood–brain barrier: The role of nanomaterials in treating neurological diseases. Adv. Mater. 2018, 30, 1801362. [Google Scholar] [CrossRef]
- Conen, A.; Raabe, A.; Schaller, K.; Fux, C.A.; Vajkoczy, P.; Trampuz, A. Management of neurosurgical implant-associated infections. Swiss Med. Wkly. 2020, 150, w20208. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, K.; Barzegar-Fallah, A.; Banstola, A.; Rizwan, S.B.; Reynolds, J.N.J. Ultrasound-Mediated Blood–Brain Barrier Disruption for Drug Delivery: A Systematic Review of Protocols, Efficacy, and Safety Outcomes from Preclinical and Clinical Studies. Pharmaceutics 2022, 14, 833. [Google Scholar] [CrossRef] [PubMed]
- Bastiancich, C.; Danhier, P.; Préat, V.; Danhier, F. Anticancer drug-loaded hydrogels as drug delivery systems for the local treatment of glioblastoma. J. Control. Release 2016, 243, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, C.; Praça, C.; Ferreira, R.; Santos, T.; Ferreira, L.; Bernardino, L. Nanoparticle-mediated brain drug delivery: Overcoming blood–brain barrier to treat neurodegenerative diseases. J. Control. Release 2016, 235, 34–47. [Google Scholar] [CrossRef]
- Anselmo, A.C.; Mitragotri, S. Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 2019, 4, e10143. [Google Scholar] [CrossRef]
- Portioli, C.; Bovi, M.; Benati, D.; Donini, M.; Perduca, M.; Romeo, A.; Dusi, S.; Monaco, H.L.; Bentivoglio, M. Novel functionalization strategies of polymeric nanoparticles as carriers for brain medications. J. Biomed. Mater. Res. Part A 2017, 105, 847–858. [Google Scholar] [CrossRef]
- Wu, H.; Lu, H.; Xiao, W.; Yang, J.; Du, H.; Shen, Y.; Qu, H.; Jia, B.; Manna, S.K.; Ramachandran, M. Sequential targeting in crosslinking nanotheranostics for tackling the multibarriers of brain tumors. Adv. Mater. 2020, 32, 1903759. [Google Scholar] [CrossRef]
- Zhang, W.; Mehta, A.; Tong, Z.; Esser, L.; Voelcker, N.H. Development of Polymeric Nanoparticles for Blood-Brain Barrier Transfer-Strategies and Challenges. Adv. Sci. 2021, 8, 2003937. [Google Scholar] [CrossRef]
- Pulgar, V.M. Transcytosis to Cross the Blood Brain Barrier, New Advancements and Challenges. Front. Neurosci. 2019, 12, 1019. [Google Scholar] [CrossRef] [PubMed]
- Mi, P.; Cabral, H.; Kataoka, K. Ligand-Installed Nanocarriers toward Precision Therapy. Adv. Mater. 2020, 32, 1902604. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.L.; Swaminathan, S.K.; Curran, G.L.; Poduslo, J.F.; Lowe, V.J.; Li, L.; Kandimalla, K.K. Apolipoprotein A-I Crosses the Blood-Brain Barrier through Clathrin-Independent and Cholesterol-Mediated Endocytosis. J. Pharmacol. Exp. Ther. 2019, 369, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Visser, C.C.; Voorwinden, L.H.; Crommelin, D.J.A.; Danhof, M.; de Boer, A.G. Characterization and Modulation of the Transferrin Receptor on Brain Capillary Endothelial Cells. Pharm. Res. 2004, 21, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, Y.; Luo, M.; Mäkilä, E.; Day, B.W.; Voelcker, N.H.; Tong, W.Y. Effectiveness of porous silicon nanoparticle treatment at inhibiting the migration of a heterogeneous glioma cell population. J. Nanobiotechnol. 2021, 19, 60. [Google Scholar] [CrossRef] [PubMed]
- Sheykhzadeh, S.; Luo, M.; Peng, B.; White, J.; Abdalla, Y.; Tang, T.; Mäkilä, E.; Voelcker, N.H.; Tong, W.Y. Transferrin-targeted porous silicon nanoparticles reduce glioblastoma cell migration across tight extracellular space. Sci. Rep. 2020, 10, 2320. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Bak, M.; Melander, F.; Thomsen, M.S.; Burkhart, A.; Kempen, P.J.; Andresen, T.L.; Moos, T. Modulating the antibody density changes the uptake and transport at the blood-brain barrier of both transferrin receptor-targeted gold nanoparticles and liposomal cargo. J. Control. Release 2019, 295, 237–249. [Google Scholar] [CrossRef]
- Kulkarni, S.A.; Feng, S.-S. Effects of Particle Size and Surface Modification on Cellular Uptake and Biodistribution of Polymeric Nanoparticles for Drug Delivery. Pharm. Res. 2013, 30, 2512–2522. [Google Scholar] [CrossRef]
- Chen, Y.P.; Chou, C.M.; Chang, T.Y.; Ting, H.; Dembélé, J.; Chu, Y.T.; Liu, T.P.; Changou, C.A.; Liu, C.W.; Chen, C.T. Bridging Size and Charge Effects of Mesoporous Silica Nanoparticles for Crossing the Blood-Brain Barrier. Front. Chem. 2022, 10, 931584. [Google Scholar] [CrossRef]
- Meng, Q.; Meng, H.; Pan, Y.; Liu, J.; Li, J.; Qi, Y.; Huang, Y. Influence of nanoparticle size on blood–brain barrier penetration and the accumulation of anti-seizure medicines in the brain. J. Mater. Chem. B 2022, 10, 271–281. [Google Scholar] [CrossRef]
- Nowak, M.; Brown, T.D.; Graham, A.; Helgeson, M.E.; Mitragotri, S. Size, shape, and flexibility influence nanoparticle transport across brain endothelium under flow. Bioeng. Transl. Med. 2020, 5, e10153. [Google Scholar] [CrossRef] [PubMed]
- Talamini, L.; Violatto, M.B.; Cai, Q.; Monopoli, M.P.; Kantner, K.; Krpetić, Ž.; Perez-Potti, A.; Cookman, J.; Garry, D.; Silveira, C.P.; et al. Influence of Size and Shape on the Anatomical Distribution of Endotoxin-Free Gold Nanoparticles. ACS Nano 2017, 11, 5519–5529. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.; Henrich-Noack, P.; Kockentiedt, S.; Hintz, W.; Tomas, J.; Sabel, B.A. Surfactants, not size or zeta-potential influence blood–brain barrier passage of polymeric nanoparticles. Eur. J. Pharm. Biopharm. 2014, 87, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.D.; Habibi, N.; Wu, D.; Lahann, J.; Mitragotri, S. Effect of Nanoparticle Composition, Size, Shape, and Stiffness on Penetration Across the Blood–Brain Barrier. ACS Biomater. Sci. Eng. 2020, 6, 4916–4928. [Google Scholar] [CrossRef]
- Li, W.; Liu, Z.; Fontana, F.; Ding, Y.; Liu, D.; Hirvonen, J.T.; Santos, H.A. Tailoring porous silicon for biomedical applications: From drug delivery to cancer immunotherapy. Adv. Mater. 2018, 30, 1703740. [Google Scholar] [CrossRef]
- Luo, M.; Lewik, G.; Ratcliffe, J.C.; Choi, C.H.J.; Mäkilä, E.; Tong, W.Y.; Voelcker, N.H. Systematic Evaluation of Transferrin-Modified Porous Silicon Nanoparticles for Targeted Delivery of Doxorubicin to Glioblastoma. ACS Appl. Mater. Interfaces 2019, 11, 33637–33649. [Google Scholar] [CrossRef]
- Zhang, D.-X.; Tieu, T.; Esser, L.; Wojnilowicz, M.; Lee, C.-H.; Cifuentes-Rius, A.; Thissen, H.; Voelcker, N.H. Differential Surface Engineering Generates Core–Shell Porous Silicon Nanoparticles for Controlled and Targeted Delivery of an Anticancer Drug. ACS Appl. Mater. Interfaces 2022, 14, 54539–54549. [Google Scholar] [CrossRef]
- Cortés-Ríos, J.; Zárate, A.M.; Figueroa, J.D.; Medina, J.; Fuentes-Lemus, E.; Rodríguez-Fernández, M.; Aliaga, M.; López-Alarcón, C. Protein quantification by bicinchoninic acid (BCA) assay follows complex kinetics and can be performed at short incubation times. Anal. Biochem. 2020, 608, 113904. [Google Scholar] [CrossRef]
- Weksler, B.; Subileau, E.; Perriere, N.; Charneau, P.; Holloway, K.; Leveque, M.; Tricoire-Leignel, H.; Nicotra, A.; Bourdoulous, S.; Turowski, P. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005, 19, 1872–1874. [Google Scholar] [CrossRef]
- Peng, B.; Tong, Z.; Tong, W.Y.; Pasic, P.J.; Oddo, A.; Dai, Y.; Luo, M.; Frescene, J.; Welch, N.G.; Easton, C.D. In situ surface modification of microfluidic blood–brain-barriers for improved screening of small molecules and nanoparticles. ACS Appl. Mater. Interfaces 2020, 12, 56753–56766. [Google Scholar] [CrossRef]
- Hubatsch, I.; Ragnarsson, E.G.; Artursson, P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nat. Protoc. 2007, 2, 2111–2119. [Google Scholar] [CrossRef]
- Bimbo, L.M.; Mäkilä, E.M.; Raula, J.; Laaksonen, T.; Laaksonen, P.; Strommer, K.; Kauppinen, E.I.; Salonen, J.J.; Linder, M.B.; Hirvonen, J.; et al. Functional hydrophobin-coating of thermally hydrocarbonized porous silicon microparticles. Biomaterials 2011, 32, 9089–9099. [Google Scholar] [CrossRef] [PubMed]
- Riikonen, J.; Nissinen, T.; Alanne, A.; Thapa, R.; Fioux, P.; Bonne, M.; Rigolet, S.; Morlet-Savary, F.; Aussenac, F.; Marichal, C.; et al. Stable surface functionalization of carbonized mesoporous silicon. Inorg. Chem. Front. 2020, 7, 631–641. [Google Scholar] [CrossRef]
- Chang, J.; Jallouli, Y.; Kroubi, M.; Yuan, X.-B.; Feng, W.; Kang, C.-S.; Pu, P.-Y.; Betbeder, D. Characterization of endocytosis of transferrin-coated PLGA nanoparticles by the blood–brain barrier. Int. J. Pharm. 2009, 379, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, R.; Deng, H.; Li, Y.; Cui, Y.; Zhang, H.; Dai, W.; He, B.; Zheng, Y.; Wang, X.; et al. Receptor mediated transcytosis in biological barrier: The influence of receptor character and their ligand density on the transmembrane pathway of active-targeting nanocarriers. Biomaterials 2018, 180, 78–90. [Google Scholar] [CrossRef]
- Li, G.; Simon, M.J.; Cancel, L.M.; Shi, Z.-D.; Ji, X.; Tarbell, J.M.; Morrison, B.; Fu, B.M. Permeability of endothelial and astrocyte cocultures: In vitro blood–brain barrier models for drug delivery studies. Ann. Biomed. Eng. 2010, 38, 2499–2511. [Google Scholar] [CrossRef]
- Yu, Y.J.; Zhang, Y.; Kenrick, M.; Hoyte, K.; Luk, W.; Lu, Y.; Atwal, J.; Elliott, J.M.; Prabhu, S.; Watts, R.J.; et al. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 2011, 3, 84ra44. [Google Scholar] [CrossRef]
- Niewoehner, J.; Bohrmann, B.; Collin, L.; Urich, E.; Sade, H.; Maier, P.; Rueger, P.; Stracke, J.O.; Lau, W.; Tissot, A.C.; et al. Increased brain penetration and potency of a therapeutic antibody using a monovalent molecular shuttle. Neuron 2014, 81, 49–60. [Google Scholar] [CrossRef]
- Wiley, D.T.; Webster, P.; Gale, A.; Davis, M.E. Transcytosis and brain uptake of transferrin-containing nanoparticles by tuning avidity to transferrin receptor. Proc. Natl. Acad. Sci. USA 2013, 110, 8662–8667. [Google Scholar] [CrossRef]
- Betzer, O.; Shilo, M.; Opochinsky, R.; Barnoy, E.; Motiei, M.; Okun, E.; Yadid, G.; Popovtzer, R. The effect of nanoparticle size on the ability to cross the blood–brain barrier: An in vivo study. Nanomedicine 2017, 12, 1533–1546. [Google Scholar] [CrossRef]
- Davies, P.F. Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nat. Clin. Pract. Cardiovasc. Med. 2009, 6, 16–26. [Google Scholar] [CrossRef]
- Dessalles, C.A.; Leclech, C.; Castagnino, A.; Barakat, A.I. Integration of substrate- and flow-derived stresses in endothelial cell mechanobiology. Commun. Biol. 2021, 4, 764. [Google Scholar] [CrossRef]
- Shurbaji, S.; Anlar, G.G.; Hussein, E.A.; Elzatahry, A.; Yalcin, H.C. Effect of Flow-Induced Shear Stress in Nanomaterial Uptake by Cells: Focus on Targeted Anti-Cancer Therapy. Cancers 2020, 12, 1916. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Sabnis, A.; Kona, S.; Nattama, S.; Patel, H.; Dong, J.-F.; Nguyen, K.T. Shear-regulated uptake of nanoparticles by endothelial cells and development of endothelial-targeting nanoparticles. J. Biomed. Mater. Res. Part A 2010, 93, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Zukerman, H.; Khoury, M.; Shammay, Y.; Sznitman, J.; Lotan, N.; Korin, N. Targeting functionalized nanoparticles to activated endothelial cells under high wall shear stress. Bioeng. Transl. Med. 2020, 5, e10151. [Google Scholar] [CrossRef] [PubMed]
- Zern, B.J.; Chacko, A.-M.; Liu, J.; Greineder, C.F.; Blankemeyer, E.R.; Radhakrishnan, R.; Muzykantov, V. Reduction of Nanoparticle Avidity Enhances the Selectivity of Vascular Targeting and PET Detection of Pulmonary Inflammation. ACS Nano 2013, 7, 2461–2469. [Google Scholar] [CrossRef] [PubMed]
- Doshi, N.; Prabhakarpandian, B.; Rea-Ramsey, A.; Pant, K.; Sundaram, S.; Mitragotri, S. Flow and adhesion of drug carriers in blood vessels depend on their shape: A study using model synthetic microvascular networks. J. Control. Release 2010, 146, 196–200. [Google Scholar] [CrossRef]
- Tan, J.; Shah, S.; Thomas, A.; Ou-Yang, H.D.; Liu, Y. The influence of size, shape and vessel geometry on nanoparticle distribution. Microfluid. Nanofluidics 2013, 14, 77–87. [Google Scholar] [CrossRef]
- Cooley, M.; Sarode, A.; Hoore, M.; Fedosov, D.A.; Mitragotri, S.; Sen Gupta, A. Influence of particle size and shape on their margination and wall-adhesion: Implications in drug delivery vehicle design across nano-to-micro scale. Nanoscale 2018, 10, 15350–15364. [Google Scholar] [CrossRef]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef]
- Williams-Medina, A.; Deblock, M.; Janigro, D. In vitro Models of the Blood-Brain Barrier: Tools in Translational Medicine. Front. Med. Technol. 2020, 2, 623950. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Zhu, D.; Tong, Z.; Peng, B.; Cheng, X.; Esser, L.; Voelcker, N.H. Influence of Surface Ligand Density and Particle Size on the Penetration of the Blood–Brain Barrier by Porous Silicon Nanoparticles. Pharmaceutics 2023, 15, 2271. https://doi.org/10.3390/pharmaceutics15092271
Zhang W, Zhu D, Tong Z, Peng B, Cheng X, Esser L, Voelcker NH. Influence of Surface Ligand Density and Particle Size on the Penetration of the Blood–Brain Barrier by Porous Silicon Nanoparticles. Pharmaceutics. 2023; 15(9):2271. https://doi.org/10.3390/pharmaceutics15092271
Chicago/Turabian StyleZhang, Weisen, Douer Zhu, Ziqiu Tong, Bo Peng, Xuan Cheng, Lars Esser, and Nicolas H. Voelcker. 2023. "Influence of Surface Ligand Density and Particle Size on the Penetration of the Blood–Brain Barrier by Porous Silicon Nanoparticles" Pharmaceutics 15, no. 9: 2271. https://doi.org/10.3390/pharmaceutics15092271
APA StyleZhang, W., Zhu, D., Tong, Z., Peng, B., Cheng, X., Esser, L., & Voelcker, N. H. (2023). Influence of Surface Ligand Density and Particle Size on the Penetration of the Blood–Brain Barrier by Porous Silicon Nanoparticles. Pharmaceutics, 15(9), 2271. https://doi.org/10.3390/pharmaceutics15092271