Screening for Methylated Poly(⌊-histidine) with Various Dimethylimidazolium/Methylimidazole/Imidazole Contents as DNA Carrier
Abstract
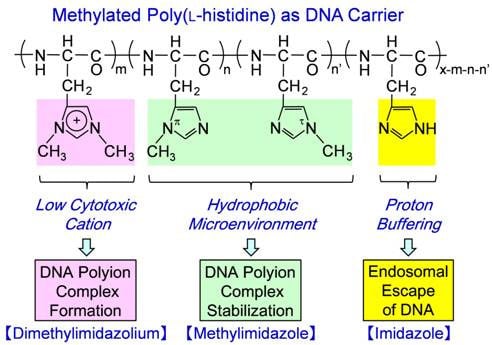
:1. Introduction

2. Experimental Section
2.1. Materials
2.2. Agarose Gel Retardation Assay
2.3. Particle Size and Zeta Potential Measurement
2.4. Transfection Procedure
3. Results and Discussion
3.1. Stability of PLH-Me/DNA Complexes

| Carrier | +/− | Particle diameter/nm | ζ Potential/mV |
|---|---|---|---|
| PLH-Me(25) | 4 | 65 ± 17 | +17 |
| 8 | 80 ± 21 | +16 | |
| 32 | 138 ± 101 | +25 | |
| PLH-Me(68) | 4 | 65 ± 18 | +6.5 |
| 8 | 67 ± 23 | +14 | |
| 32 | 123 ± 47 | +19 | |
| PLH-Me(87) | 4 | 82 ± 22 | +26 |
| 8 | 51 ± 15 | +23 | |
| 32 | 61 ± 19 | +21 |
3.2. Gene Delivery by PLH-Me/DNA Complexes

4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Itaka, K.; Ishii, T.; Hasegawa, Y.; Kataoka, K. Biodegradable polyamino acid-based polycations as safe and effective gene carrier minimizing cumulative toxicity. Biomaterials 2010, 31, 3707–3714. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Oba, M.; Nakanishi, M.; Fukushima, S.; Yamasaki, Y.; Koyama, H.; Nishiyama, N.; Kataoka, K. Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J. Am. Chem. Soc. 2008, 130, 16287–16294. [Google Scholar] [CrossRef] [PubMed]
- Pouton, L.W.; Seymour, L.W. Key issues in non-viral gene delivery. Adv. Drug. Deliver. Rev. 2001, 46, 187–203. [Google Scholar] [CrossRef]
- Steinman, R.M.; Mellman, I.S.; Muller, W.A.; Cohn, Z.A. Endocytosis and the recycling of plasma membrane. J. Cell Biol. 1983, 96, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Margny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed]
- Midoux, P.; Pichon, C.; Yaouanc, J.J.; Jaffrès, P.A. Chemical vectors for gene delivery: A current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 2009, 157, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Ihm, J.E.; Han, K.O.; Han, I.K.; Ahn, K.D.; Han, D.K.; Cho, C.S. High transfection efficiency of poly(4-vinylimidazole) as a new gene carrier. Bioconjugate Chem. 2003, 14, 707–708. [Google Scholar] [CrossRef] [PubMed]
- Roufaï, M.B.; Midoux, P. Histidylated polylysine as DNA vector: Elevation of the imidazole protonation and reduced cellular uptake without change in the polyfection efficiency of serum stabilized negative polyplexes. Bioconjugate Chem. 2001, 12, 92–99. [Google Scholar] [CrossRef]
- Mével, M.; Neveu, C.; Gonçalves, C.; Yaouanc, J.J.; Pichon, C.; Jaffrès, P.A.; Midoux, P. Novel neutral imidazole-lipophosphoramides for transfection assays. Chem. Commun. 2008. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Matsuda, K.; Negishi, Y.; Kawakami, H. Intracellular co-delivery of zinc ions and plasmid DNA for enhancing gene transfection activity. Metallomics 2014, 6, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Nishinohara, S.; Kawakami, H. Zinc-chelated poly(l-vinylimidazole) and a carbohydrate ligand polycation form DNA ternary complexes for gene delivery. Bioconjugate Chem. 2011, 22, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Hakamatani, T.; Kawakami, H. Synthesis and characterization of alkylated poly(1-vinylimidazole) to control the stability of its DNA polyion complexes for gene delivery. Bioconjugate Chem. 2010, 21, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Sudo, M.; Nagaoka, S.; Kawakami, H. Carboxymethyl poly(l-histidine) as a new pH-sensitive polypeptide to enhance polyplex gene delivery. Mol. Pharm. 2008, 5, 898–901. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Sekine, T.; Kawakami, H.; Nagaoka, S. Design of aminated poly(1-vinylimidazole) for a new pH-sensitive polycation to enhance cell-specific gene delivery. Bioconjugate Chem. 2007, 18, 1662–1667. [Google Scholar] [CrossRef] [PubMed]
- Mével, M.; Breuzard, G.; Yaouanc, J.J.; Clément, J.C.; Lehn, P.; Pichon, C.; Jaffrès, P.A.; Midoux, P. Synthesis and transfection activity of new cationic phosphoramidate lipids: high efficiency of an imidazolium derivative. Chem. Bio. Chem. 2008, 9, 1462–1471. [Google Scholar] [CrossRef] [PubMed]
- Perche, F.; Benvegnu, T.; Berchel, M.; Lebegue, L.; Pichon, C.; Jaffrès, P.A.; Midoux, P. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine 2011, 7, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, C.; Berchel, M.; Gosselin, M.P.; Malard, V.; Cheradame, H.; Jaffrès, P.A.; Guégan, P.; Pichon, C.; Midoux, P. Lipopolyplexes comprising imidazole/imidazolium lipophosphoramidate, histidinylated polyethyleneimine and siRNA as efficient formulation for siRNA transfection. Int. J. Pharm. 2014, 460, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Delalande, A.; Gosselin, M.P.; Suwalski, A.; Guilmain, W.; Leduc, C.; Berchel, M.; Jaffrès, P.A.; Baril, P.; Midoux, P.; Pichon, C. Enhanced Achilles tendon healing by fibromodulin gene transfer. Nanomedicine 2015. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Kumagai, T.; Kawakami, H. Synthesis and characterization of methylated poly(l-histidine) to control the stability of its siRNA polyion complexes for RNAi. Bioconjugate Chem. 2012, 23, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Zamenhof, S. Preparation and assay of deoxyribonucleic acid from animal tissue. Methods Enzymol. 1957, 3, 696–704. [Google Scholar]
- Chittimalla, C.; Zammut-Italiano, L.; Zuber, G.; Behr, J.P. Monomolecular DNA nanoparticles for intravenous delivery of genes. J. Am. Chem. Soc. 2005, 127, 11436–11441. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asayama, S.; Kumagai, T.; Kawakami, H. Screening for Methylated Poly(⌊-histidine) with Various Dimethylimidazolium/Methylimidazole/Imidazole Contents as DNA Carrier. Pharmaceutics 2015, 7, 224-232. https://doi.org/10.3390/pharmaceutics7030224
Asayama S, Kumagai T, Kawakami H. Screening for Methylated Poly(⌊-histidine) with Various Dimethylimidazolium/Methylimidazole/Imidazole Contents as DNA Carrier. Pharmaceutics. 2015; 7(3):224-232. https://doi.org/10.3390/pharmaceutics7030224
Chicago/Turabian StyleAsayama, Shoichiro, Takao Kumagai, and Hiroyoshi Kawakami. 2015. "Screening for Methylated Poly(⌊-histidine) with Various Dimethylimidazolium/Methylimidazole/Imidazole Contents as DNA Carrier" Pharmaceutics 7, no. 3: 224-232. https://doi.org/10.3390/pharmaceutics7030224
APA StyleAsayama, S., Kumagai, T., & Kawakami, H. (2015). Screening for Methylated Poly(⌊-histidine) with Various Dimethylimidazolium/Methylimidazole/Imidazole Contents as DNA Carrier. Pharmaceutics, 7(3), 224-232. https://doi.org/10.3390/pharmaceutics7030224