A Hybrid Robust-Learning Architecture for Medical Image Segmentation with Noisy Labels
Abstract
:1. Introduction
- (1)
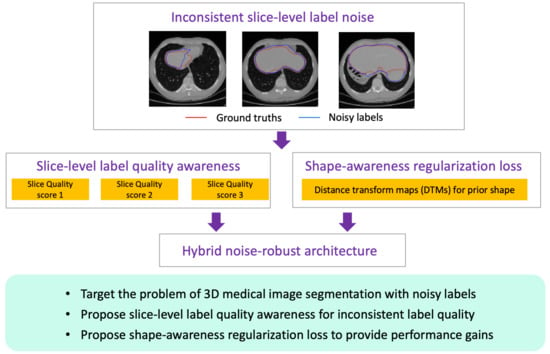
- Different from previous studies, we target the challenging problem of 3D medical image segmentation with noisy labels, especially the inconsistent noisy label qualities among different slices. To address this problem, we propose a novel end-to-end hybrid robust-learning architecture to combat noisy labels from the perspective of slice-level label-quality awareness;
- (2)
- We propose a novel slice-level label-quality awareness method, which automatically generates quality scores for each slice in a set without knowing the prior noise distribution. With the help of re-weighting, our method can alleviate the negative effect of noisy labels. The design is particularly effective for 3D medical image segmentation by satisfying the constraints of noise tolerance and the capacity limitations of GPUs;
- (3)
- We propose a shape-awareness regularization loss to introduce prior shape information to provide extra performance gains. In the presence of noisy labels, we regard it as an auxiliary loss instead of the main learning targets and, further, it benefits the model training together with slice-level label-quality awareness. To our knowledge, this is the first attempt to apply prior shape information for the problem of learning with noisy labels.
2. Related Works
3. Methods
3.1. Segmentation Module
3.2. Label-Quality Awareness Module
3.3. Shape-Awareness Regularization Loss
3.4. The Final Framework
4. Experiments and Results
4.1. Data and Implementation Details
4.2. Comparisons on Liver Segmentation Dataset
4.3. Comparisons on Multi-Organ Segmentation Dataset
4.4. Ablation Study and Visualization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, H.; Liu, X.; Sun, S.; Yan, X.; Xie, X. Recurrent mask refinement for few-shot medical image segmentation. arXiv 2021, arXiv:2108.00622. [Google Scholar]
- Liu, L.; Cheng, J.; Quan, Q.; Wu, F.-X.; Wang, Y.-Y.; Wang, J. A survey on U-shaped networks in medical image segmentations. Neurocomputing 2020, 409, 244–258. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A hybrid transformer architecture for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Strasbourg, France, 27 September–1 October 2021. [Google Scholar]
- Xue, Y.; Tang, H.; Qiao, Z.; Gong, G.; Yin, Y.; Qian, Z.; Huang, C.; Fan, W.; Huang, X. Shape-aware organ segmentation by predicting signed distance maps. In Proceedings of the 34th AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 12565–12572. [Google Scholar]
- Patrini, G.; Rozza, A.; Krishna Menon, A.; Nock, R.; Qu, L. Making Deep Neural Networks Robust to Label Noise: A Loss Correction Approach. arXiv 2017, arXiv:1609.03683. [Google Scholar]
- Hendrycks, D.; Mazeika, M.; Wilson, D.; Gimpel, K. Using trusted data to train deep networks on labels corrupted by severe noise. arXiv 2018, arXiv:1802.05300. [Google Scholar]
- Wang, Z.; Hu, G.; Hu, Q. Training noise-robust deep neural networks via meta-learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- Dgani, Y.; Greenspan, H.; Goldberger, J. Training a neural network based on unreliable human annotation of medical images. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI), Washington, DC, USA, 4–7 April 2018; pp. 39–42. [Google Scholar]
- Zhang, Z.; Sabuncu, M.R. Generalized cross entropy loss for training deep neural networks with noisy labels. In Proceedings of the 32nd Conference on Neural Information Processing Systems (NeurIPS), Montréal, QC, Canada, 3–8 December 2018. [Google Scholar]
- Wang, Y.; Ma, X.; Chen, Z.; Luo, Y.; Yi, J.; Bailey, J. Symmetric cross entropy for robust learning with noisy labels. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019. pp. 322–330.
- Karimi, D.; Dou, H.; Warfield, S.K.; Gholipour, A. Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis. Med. Image Anal. 2020, 65, 101759. [Google Scholar] [CrossRef] [PubMed]
- Mirikharaji, Z.; Yan, Y.; Hamarneh, G. Learning to segment skin lesions from noisy annotations. arXiv 2019, arXiv:1906.03815. [Google Scholar]
- Ren, M.; Zeng, W.; Yang, B.; Urtasun, R. Learning to reweight examples for robust deep learning. In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 4334–4343. [Google Scholar]
- Zhu, H.; Shi, J.; Wu, J. Pick-and-learn: Automatic quality evaluation for noisy-labeled image segmentation. arXiv 2019, arXiv:1907.11835v1. [Google Scholar]
- Zhang, T.; Yu, L.; Hu, N.; Lv, S.; Gu, S. Robust medical image segmentation from non-expert annotations with tri-network. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020, 23rd International Conference, Lima, Peru, 4–8 October 2020; pp. 249–258. [Google Scholar]
- Han, B.; Yao, Q.; Yu, X.; Niu, G.; Xu, M.; Hu, W.; Tsang, I.; Sugiyama, M. Co-teaching: Robust training of deep neural networks with extremely noisy label. In Proceedings of the Conference on Neural Information Processing Systems (NeurIPS), Montréal, QC, Canada, 3–8 December 2018. [Google Scholar]
- Song, H.; Kim, M.; Park, D.; Shin, Y.; Lee, J.-G. Learning from noisy labels with deep neural networks: A survey. arXiv 2020, arXiv:2007.08199. [Google Scholar]
- Goldberger, J.; Ben-Reuven, E. Training deep neural-networks using a noise adaptation layer. In Proceedings of the 4th International Conference on Learning Representations (ICLR 2016), San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Han, B.; Yao, J.; Niu, G.; Zhou, M.; Tsang, I.; Zhang, Y.; Sugiyama, M. Masking: A new perspective of noisy supervision. In Proceedings of the 32nd Conference on Neural Information Processing Systems (NeurIPS), Montréal, QC, Canada, 3–8 December 2018. [Google Scholar]
- Ghosh, A.; Kumar, H.; Sastry, P. Robust loss functions under label noise for deep neural networks. In Proceedings of the 31st AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Menon, A.K.; Rawat, A.S.; Reddi, S.J.; Kumar, S. Can gradient clipping mitigate label noise? In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Liu, S.; Niles-Weed, J.; Razavian, N.; Fernandez-Granda, C. Early-learning regularization prevents memorization of noisy labels. In Proceedings of the Advances in Neural Information Processing Systems 33: Annual Conference on Neural Information Processing Systems (NeurIPS 2020), Virtual, 6–12 December 2020. [Google Scholar]
- Tanno, R.; Saeedi, A.; Sankaranarayanan, S.; Alexander, D.C.; Silberman, N. Learning from noisy labels by regularized estimation of annotator confusion. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019. [Google Scholar]
- Jindal, I.; Nokleby, M.; Chen, X. Learning deep networks from noisy labels with dropout regularization. In Proceedings of the 2016 IEEE 16th International Conference on Data Mining (ICDM), Barcelona, Spain, 12–15 December 2016. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. Mixup: Beyond empirical risk minimization. In Proceedings of the 5th International Conference on Learning Representations (ICLR), Toulon, France, 24–26 April 2017. [Google Scholar]
- Nishi, K.; Ding, Y.; Rich, A.; Höllerer, T. Augmentation strategies for learning with noisy labels. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021. [Google Scholar]
- Zheltonozhskii, E.; Baskin, C.; Mendelson, A.; Bronstein, A.M.; Litany, O. Contrast to divide: Self-supervised pre-training for learning with noisy labels. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 4–8 January 2022. [Google Scholar]
- Jiang, L.; Zhou, Z.; Leung, T.; Li, L.-J.; Li, F.-F. Mentornet: Learning data-driven curriculum for very deep neural networks on corrupted labels. In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018. [Google Scholar]
- Arazo, E.; Ortego, D.; Albert, P.; O’Connor, N.E.; McGuinness, K. Unsupervised label noise modeling and loss correction. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Han, J.; Luo, P.; Wang, X. Deep self-learning from noisy labels. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019. [Google Scholar]
- Lee, K.-H.; He, X.; Zhang, L.; Yang, L. Cleannet: Transfer learning for scalable image classifier training with label noise. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Shu, J.; Xie, Q.; Yi, L.; Zhao, Q.; Zhou, S.; Xu, Z.; Meng, D. Meta-weight-net: Learning an explicit mapping for sample weighting. In Proceedings of the Conference on Neural Information Processing Systems (NeurIPS), Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Min, S.; Chen, X.; Zha, Z.J.; Wu, F.; Zhang, Y. A two-stream mutual attention network for semi-supervised biomedical segmentation with noisy labels. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 4578–4585. [Google Scholar]
- Malach, E.; Shalev-Shwartz, S. Decoupling “when to update” from “how to update”. In Proceedings of the 31st Conference on Neural Information Processing Systems (NeurIPS), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the 19th International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Xu, Y.; Zhu, L.; Jiang, L.; Yang, Y. Faster meta update strategy for noise-robust deep learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 144–153. [Google Scholar]
- Han, B.; Yao, Q.; Liu, T.; Niu, G.; Tsang, I.W.; Kwok, J.T.; Sugiyama, M. A survey of label-noise representation learning: Past, present and future. arXiv 2020, arXiv:2011.04406. [Google Scholar]
- Zheng, H.; Zhang, Y.; Yang, L.; Liang, P.; Zhao, Z.; Wang, C.; Chen, D.Z. A new ensemble learning framework for 3D biomedical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; pp. 5909–5916. [Google Scholar]
- Ma, J.; Wei, Z.; Zhang, Y.; Wang, Y.; Lv, R.; Zhu, C.; Gaoxiang, C.; Liu, J.; Peng, C.; Wang, L.; et al. How distance transform maps boost segmentation CNNs: An empirical study. In Proceedings of the Medical Imaging with Deep Learning, Montreal, QC, Canada, 6–9 July 2020; pp. 479–492. [Google Scholar]
- Karimi, D.; Salcudean, S.E. Reducing the hausdorff distance in medical image segmentation with convolutional neural networks. IEEE Trans. Med. Imaging 2019, 39, 499–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaos Challenge. Available online: https://doi.org/10.5281/zenodo.3431873 (accessed on 10 January 2022).
- Shi, J.; Ding, X.; Liu, X.; Li, Y.; Liang, W.; Wu, J. Automatic clinical target volume delineation for cervical cancer in CT images using deep learning. Med. Phys. 2021, 48, 3968–3981. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wu, J. Distilling effective supervision for robust medical image segmentation with noisy labels. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021. [Google Scholar]




| Method | Noise Level 1 | Noise Level 2 | ||||
|---|---|---|---|---|---|---|
| 25% | 50% | 75% | 25% | 50% | 75% | |
| Plain [35] | 70.94 ± 0.96 | 68.97 ± 0.43 | 65.24 ± 1.32 | 59.61 ± 0.41 | 56.82 ± 0.19 | 48.07 ± 2.38 |
| Pick-and-learn [14] | 65.67 ± 0.44 | 59.36 ± 0.73 | 54.10 ± 0.86 | 50.91 ± 0.59 | 47.31 ± 0.26 | 40.32 ± 0.45 |
| Disagreement [33] | 71.43 ± 1.10 | 69.88 ± 0.24 | 67.58 ± 0.28 | 66.64 ± 0.07 | 55.72 ± 0.19 | 47.23 ± 0.25 |
| INT [45] | 77.34 ± 0.14 | 75.33 ± 0.08 | 71.67 ± 0.06 | 70.38 ± 0.81 | 60.80 ± 0.64 | 53.56 ± 0.67 |
| Area-aware [44] | 76.62 ± 1.75 | 74.92 ± 1.27 | 69.45 ± 1.96 | 70.63 ± 1.32 | 60.44 ± 1.98 | 54.82 ± 1.56 |
| Ours | 78.31 ± 0.46 | 76.29 ± 0.63 | 72.78 ± 0.60 | 71.72 ± 0.18 | 64.05 ± 0.30 | 56.99 ± 0.66 |
| Noise Rates | Method | Liver | Right Kidney | Left Kidney | Spleen | Average |
|---|---|---|---|---|---|---|
| No noise | Plain [35] | 84.20 | 75.13 | 64.93 | 73.66 | 74.48 |
| Plain [35] | 79.36 | 55.09 | 42.88 | 52.51 | 57.46 | |
| Pick-and-learn [14] | 80.31 | 45.79 | 38.33 | 38.44 | 50.72 | |
| 25% | Disagreement [33] | 73.46 | 49.79 | 44.66 | 52.09 | 55.00 |
| INT [45] | 78.93 | 56.63 | 45.66 | 59.14 | 60.09 | |
| Area-aware [44] | 75.05 | 52.83 | 54.17 | 51.11 | 58.29 | |
| Ours | 78.00 | 60.72 | 49.62 | 60.95 | 62.32 | |
| Plain [35] | 76.86 | 52.43 | 42.75 | 54.40 | 56.61 | |
| Pick-and-learn [14] | 70.36 | 48.87 | 41.26 | 48.55 | 52.26 | |
| 50% | Disagreement [33] | 71.37 | 49.87 | 41.26 | 49.55 | 53.01 |
| INT [45] | 79.10 | 55.10 | 46.97 | 56.29 | 59.37 | |
| Area-aware [44] | 75.47 | 51.80 | 46.66 | 54.19 | 57.03 | |
| Ours | 80.27 | 54.07 | 47.54 | 60.49 | 61.60 | |
| Plain [35] | 75.18 | 56.29 | 41.75 | 52.10 | 56.33 | |
| Pick-and-learn [14] | 70.79 | 48.13 | 36.83 | 44.22 | 49.99 | |
| 75% | Disagreement [33] | 72.99 | 48.49 | 40.41 | 46.42 | 52.08 |
| INT [45] | 76.66 | 50.69 | 47.25 | 57.63 | 58.06 | |
| Area-aware [44] | 76.47 | 48.99 | 45.12 | 57.20 | 56.94 | |
| Ours | 78.62 | 52.08 | 51.84 | 59.62 | 60.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Guo, C.; Wu, J. A Hybrid Robust-Learning Architecture for Medical Image Segmentation with Noisy Labels. Future Internet 2022, 14, 41. https://doi.org/10.3390/fi14020041
Shi J, Guo C, Wu J. A Hybrid Robust-Learning Architecture for Medical Image Segmentation with Noisy Labels. Future Internet. 2022; 14(2):41. https://doi.org/10.3390/fi14020041
Chicago/Turabian StyleShi, Jialin, Chenyi Guo, and Ji Wu. 2022. "A Hybrid Robust-Learning Architecture for Medical Image Segmentation with Noisy Labels" Future Internet 14, no. 2: 41. https://doi.org/10.3390/fi14020041
APA StyleShi, J., Guo, C., & Wu, J. (2022). A Hybrid Robust-Learning Architecture for Medical Image Segmentation with Noisy Labels. Future Internet, 14(2), 41. https://doi.org/10.3390/fi14020041